How CLIJ2 can make your bio-image analysis workflows incredibly fast
Posted by Robert Haase, on 14 July 2020
Do you also spend a substantial amount of your time waiting for automated image analysis to finish? I did, again and again for more than a decade. And, then, the morning after running a script overnight, you realize that a parameter was wrongly set. It was about time to change that.
History
Processing on graphics processing units (GPUs) can be incredibly fast and the concepts behind are known for quite some time. The Open Computing Language (OpenCL) for programming GPUs from various vendors is already a decade old. However, only experts speak that language. It also took imaging scientists quite a while until they adapted it and started exploiting GPUs for specific purposes such as deconvolution. The end users still didn’t have access to the full power of general-purpose GPU-acceleration. Specifically, in the ImageJ or Fiji platform, an easy way to use collection of GPU-accelerated image processing operations was missing. When I joined Gene Myers lab, Loic Royer, Uwe Schmidt, Martin Weigert and Alexandr Dibrov had already started collecting such GPU-accelerated operations in OpenCL, for example for applying online real-time multi-view image fusion on one of the light-sheet microscopes in the lab. I was totally amazed by the achievements of my colleagues and how fast image processing can be done on GPUs (Movie 1). Everyone heard about it. We were told during computer science lectures. But I never saw it in action in the context of bio-imaging analysis.
By chance, that was about the time when I made auto-completion run in Fijis script editor because I wanted to simplify building workflows in ImageJ in general. Users world-wide sent lot of positive feedback confirming ImageJ macro-scripting became much easier with this new feature. It was obvious to me that auto-completion could also pave the path towards building GPU-accelerated image analysis workflows.
From there it was one more year of work: writing about one hundred GPU-accelerated operations, detailed benchmarking, automated quality assurance, user-documentation, example workflows in five scripting languages and a website bringing all that together in an accessible way. Ultimate goal: make GPUs accessible to biologists, without the need to learn a new programming language and without the need to talk to us. Open Science.
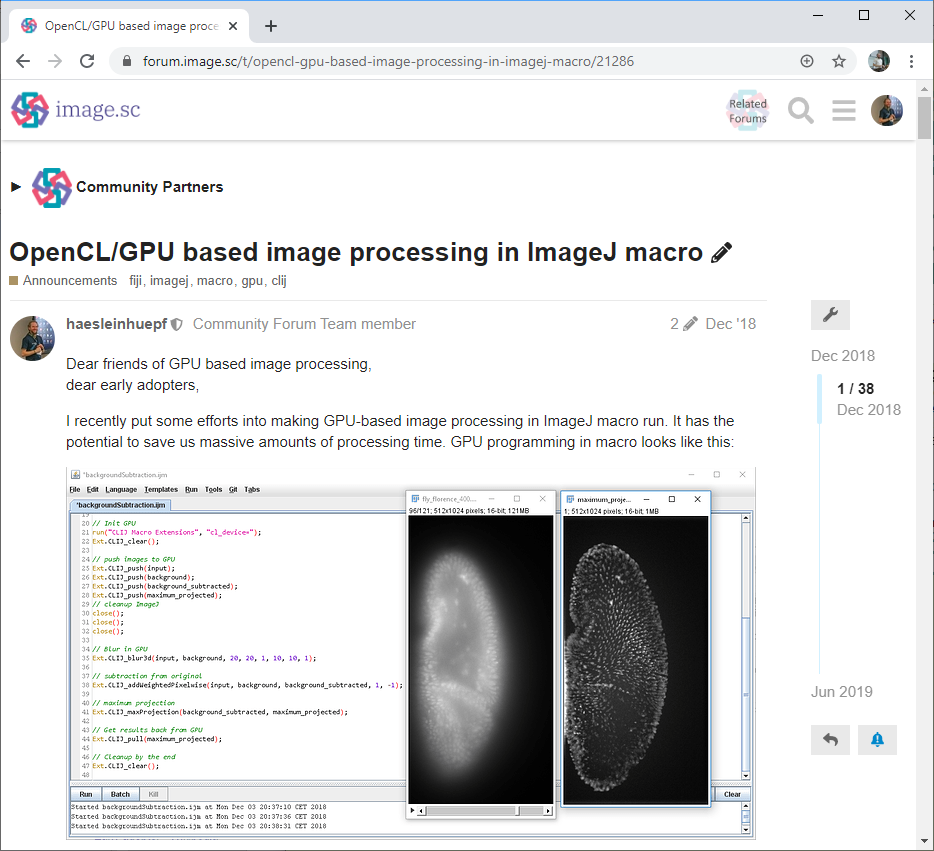
Half the way down that road, I posted on the image.sc forum asking for feedback: who would use such a bridge between OpenCL and ImageJ (we termed CLIJ) and for what? (Fig. 1) This was a fascinating moment for me as an open science advocate as it was not clear yet if CLIJ will become a thing. And it was not obvious if others were working on similar ideas. From the computer scientists’ perspective it was also risky; we cannot just build tools. We also need to publish papers about them. We could have been spoofed at that point. But it was all going right. The feedback from the community was overwhelming and CLIJ started to fly. Actually, I doubt that the rest of the project would have been possible without the community feedback. The paper with detailed benchmarking results came out about one year after the announcement on the forum.
What’s new in CLIJ2
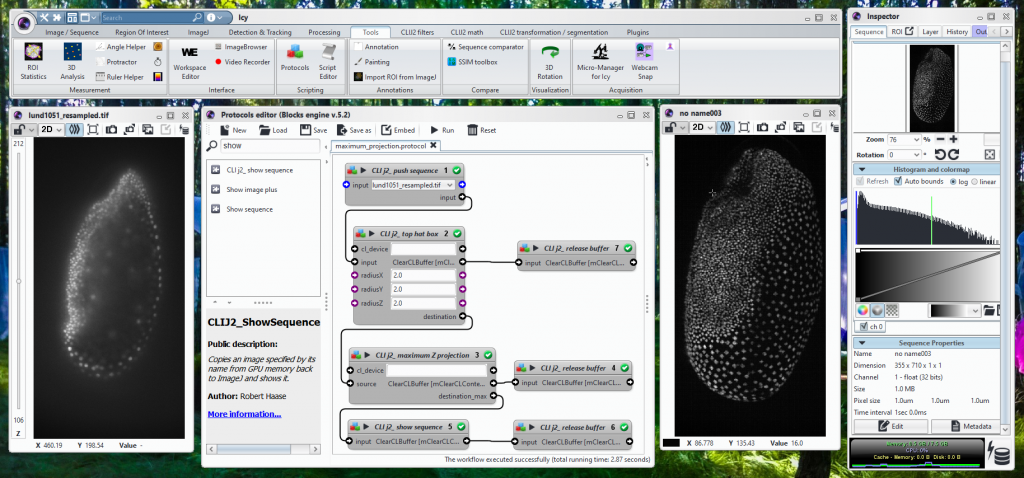
Now, a year after CLIJs release, its successor CLIJ2 is maturing, with more functionality, faster in the warmup and more detailed documentation on a new website. CLIJ2 also introduces new concepts to the ImageJ/Fiji universe: graph-based image analysis for studying relationships between adjacent cells. Again, mathematicians have known the underlying concepts and algorithms for many years, but there was no easy way for life-scientists to just use tools based on graphs representing cells in images in Fiji. Furthermore, CLIJ2 eventually supports Icy Bioimaging and Matlab. Why so? We want to build bridges between these image-analysis communities (Movie 2) (Fig. 2).
Movie 2: CLIJ2 enables studying relationships between neighboring objects such as cells in two and three dimensions with the ease-of-use known from CLIJ.
How you can exploit GPU-acceleration for your science
CLIJ is made in a way so that ImageJ users find well-known concepts when building GPU-accelerated workflows. If you can write ImageJ macros, you can write GPU-accelerated CLIJ macros. After activating the “clij” and “clij2” update sites in Fiji, all CLIJ2 operations can be found in the menu Plugins > ImageJ on the GPU (CLIJ2). When using them for the first time you may realize executing some of these operations running on a GPU may not be much faster than executing the good old analogous ImageJ/Fiji menus. The reason is that GPU-accelerated CLIJ2 plugins push the image to the GPU memory, process it there and pull the result back. Pushing and pulling takes time. If it takes more time than the operation on the CPU, it is not worth using the GPU for it. In order to really exploit the power of GPUs, one needs to implement entire workflows on the GPU and not pulling and pushing intermediate results back and forth. What sounds complicated comes at the same user convenience experienced ImageJ macro users know already: the macro recorder! (Movie 3)
Some bio-image analysis workflow designers are happy with clicking menus and using the recorder, others prefer to use the auto-completion in Fijis Script Editor. This works best in combination with the new website, which lists typical methods called after the method you were just using and points towards examples and tutorials (Movie 4).
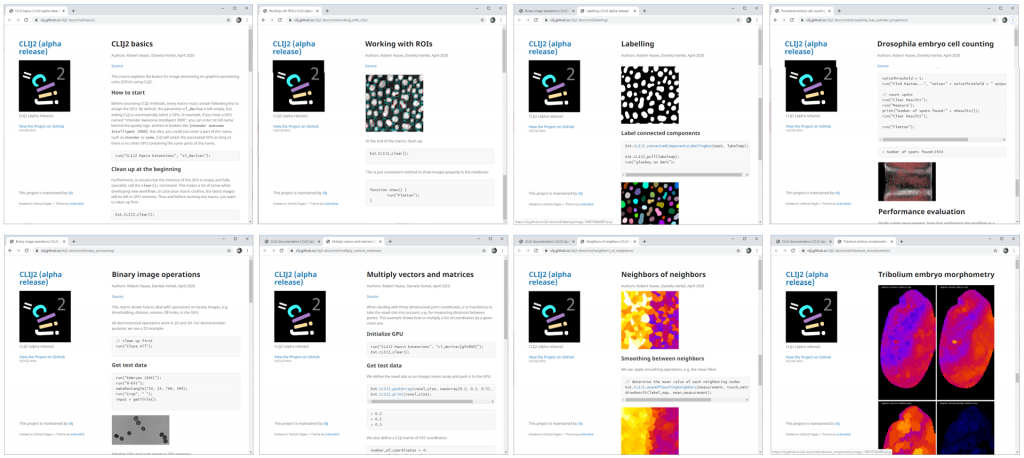
Of course, if you want to translate a workflow step-by-step to CLIJ2, you need a workflow in advance. The website provides some example workflows for typical use cases (Fig. 3).
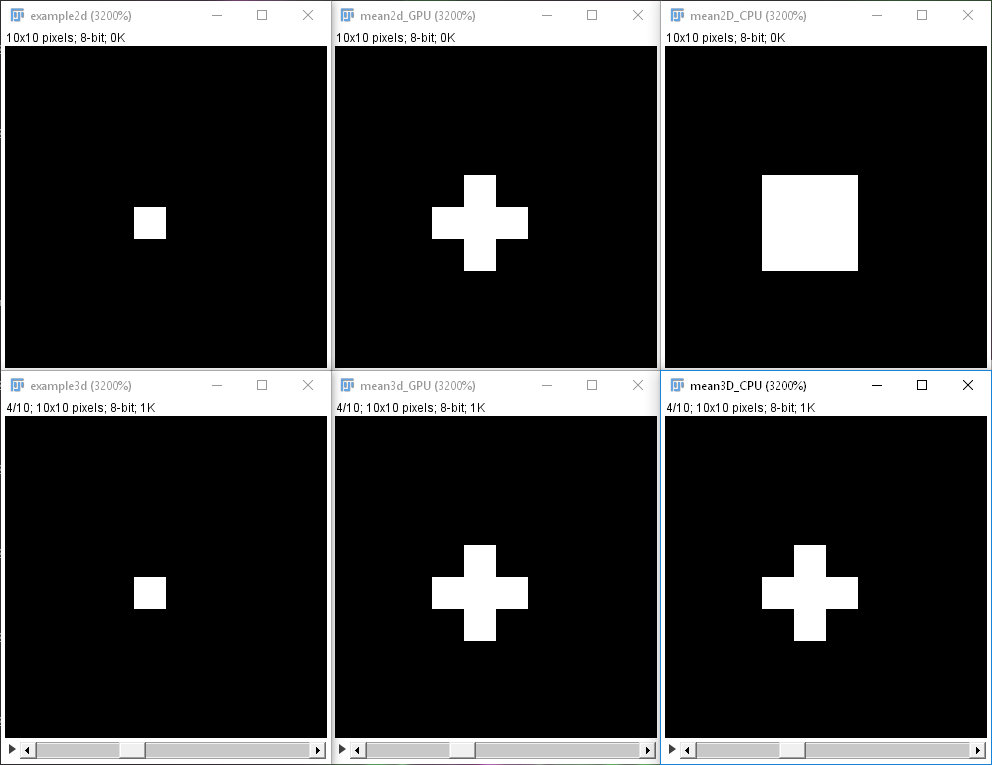
After translating your workflows, you may also want to measure how much faster it gets using CLIJ2 to learn what you can gain. Furthermore, please be thorough and check if the CLIJ2 workflow does the same as your original workflow. This shall be mentioned because not all GPU-accelerated operations do exactly the same as their ImageJ counterparts. In practice, this should be negligible, but one needs to check. Depending on the dimensionality of the dataset, Fiji considers different neighborhoods of pixels, CLIJ does not. We decided not to copy the observed discrepancy, and this is why these filters are somewhat different between ImageJ and CLIJ (Fig. 4).
Summary
Constructing CLIJ2 workflows enables new ways of processing images in ImageJ and Fiji (Fig. 5). While some new CLIJ commands are a bit more complicated than older ImageJ macro commands, the concepts for writing them, such as the recorder and auto-completion, are the same.

Learning how to use CLIJ2 is worth the effort; if you can spare hundreds of hours of processing time, it would even be worth investing a hundred hours in learning it. However, we are spending increased efforts in simplifying the transition. A website with introductory tutorials, example macros and a frequently asked questions section, cheat sheets (Fig. 6), as well as the full reference is supposed to make the life of users easy. CLIJ2 was the subject of one of the sessions recorded online for NEUBIAS academy @Home webinars. Last but not least, support is provided via image.sc. And if you have an idea of how to make GPU-accelerated image processing even easier to use, feedback is very welcome!
Acknowledgements
Development of CLIJ is a community effort. We would like to thank everybody who helped developing and testing. In particular thanks goes to Alex Herbert (University of Sussex), Bram van den Broek (Netherlands Cancer Institute), Brenton Cavanagh (RCSI), Brian Northan (True North Intelligent Algorithms), Bruno C. Vellutini (MPI CBG), Curtis Rueden (UW-Madison LOCI), Damir Krunic (DKFZ), Daniela Vorkel (MPI CBG), Daniel J. White (GE), Eduardo Conde-Sousa (University of Porto), Erick Ratamero (The Jackson Laboratory), Gaby G. Martins (IGC), Guillaume Witz (Bern University), Giovanni Cardone (MPI Biochem), Jan Brocher (Biovoxxel), Jean-Yves Tinevez (Institute Pasteur), Johannes Girstmair (MPI CBG), Juergen Gluch (Fraunhofer IKTS), Kota Miura, Laurent Thomas (Acquifer), Matthew Foley (University of Sydney), Matthias Arzt (MPI-CBG), Nico Stuurman (UCSF), Nik Cordes (Los Alamos National Laboratory), Ofra Golani (Weizmann Institute of Science), Peter Haub, Pete Bankhead (University of Edinburgh), Pit Kludig, Pradeep Rajasekhar (Monash University), Ruth Whelan-Jeans, Siân Culley (LMCB MRC), Tanner Fadero (UNC-Chapel Hill), Thomas Irmer (Zeiss), Tobias Pietzsch (MPI-CBG), Wilson Adams (VU Biophotonics).
Reusing this material
This blog post is open-access. Figures and text can be rused under the terms of the CC BY 4.0 license unless mentioned otherwise.

Robert Haase
Postdoctoral Scientists
Gene Myers’ Lab
MPI-CBG, Dresden, Germany


 (11 votes, average: 1.00 out of 1)
(11 votes, average: 1.00 out of 1)
