Etch A Cell – segmenting electron microscopy data with the power of the crowd
Posted by Martin Jones, on 16 July 2020
Recent years have seen remarkable developments in imaging techniques and technologies, producing increasingly rich datasets that require huge amounts of costly technological infrastructure, computational power and researcher effort to process. Techniques such as light-sheet microscopy and volume electron microscopy routinely generate terabytes worth of data overnight. With a single data acquisition producing more images than any single microscopist could realistically analyse in detail, vast swathes of data can be left underutilised or simply unexamined.
Automated approaches have been successfully applied to alleviate the analytical bottleneck of a number of research areas within bioimaging. However, many substantial analysis challenges remain – in electron microscopy (EM), for example, automated methods are yet to provide a robust and generalisable analysis solution for all but a handful of specific applications. Consequently, the vast majority of EM data analysis remains a stubbornly manual process. Typically, the analysis of micrographs involves the task of segmentation (delineating objects of interest from the background), and is frequently performed by an expert laboriously hand-tracing around the objects with a stylus or their mouse.
Providing a ‘shining light’ that has already revolutionised much modern image analysis is deep-learning; a subclass of machine learning that has proven highly effective at solving computer vision challenges across many domains, including microscopy [1]. Broadly speaking, deep-learning approaches have solved many previously impenetrable analysis challenges through shifting the task of complex algorithm design from the researcher to the computer itself. This enables ever-more efficient methods and advances in computational hardware to be more easily leveraged for fast and powerful analysis.
However, nothing comes for free – the analytical advances of deep-learning conceal a ‘sleight of hand’ that shifts the research bottleneck to a different aspect of the workflow, rather than eliminating it altogether. Supervised deep-learning methods require the researcher to feed the algorithm pairs of images: the raw data in conjunction with the answer they would like the system to be able to reproduce, much like a teacher giving mock exam questions and model answers to their students. Once the algorithm is fully trained (able to reliably reproduce the ‘ground truth’ answers provided), then it should be capable of providing predictions for new datasets that lack the corresponding ground truth. As a result, this moves the research bottleneck to the generation of the ground truth data required for the training of deep-learning models. As it is often necessary to provide training data that are representative of all the variations and configurations naturally present within the data, the quantities of ground truth necessary for training a robust model can be voluminous (the maxim of ‘the best data is more data’ is frequently used), and there often simply aren’t enough experts with time to provide sufficient quantities of annotations.
The challenge of scalable data analysis is not unique to EM; many other research domains have experienced similar revolutions in their data-generating capacity, while data examination has remained a slow, and often subjective, process dependent on human pattern-recognition abilities. An approach that is being increasingly applied to solve this challenge is to perform distributed data analysis through engaging the public via online citizen science. This has already been applied with much success in other fields of research, with many well-known projects particularly in astronomy (e.g. Galaxy Zoo) and ecology (e.g. Snapshot Serengeti). Inspired by this, we approached one of the largest online citizen science platforms, the Zooniverse, to explore the possibility of involving volunteers in the segmentation of EM images. These conversations led to the collaborative development of an online project, ‘Etch A Cell’, through which volunteers could trace around organelles within their web browser.
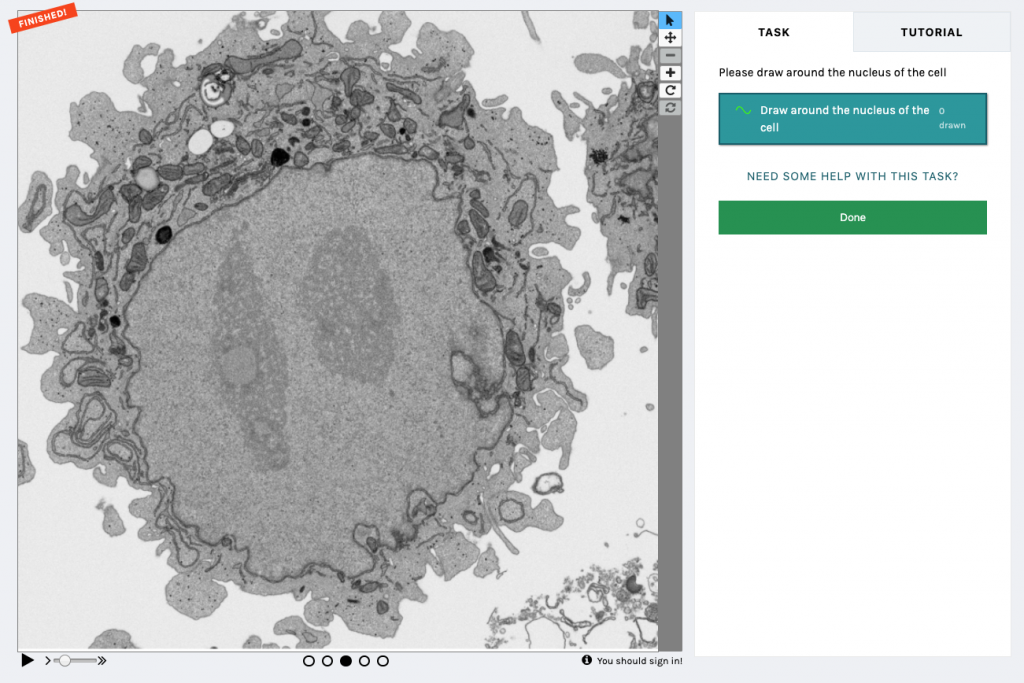
In short, the (first) Etch A Cell project asked volunteers to perform the task of nuclear envelope segmentation using a drawing tool (Fig.1) . As with all Zooniverse projects, Etch A Cell was available for anyone with an internet connection to contribute to. After accessing the classification interface for the project, volunteers were introduced to the segmentation task through a detailed Tutorial. Background information was also provided in the project interface, and a Talk forum provided a place for the volunteer community to ask further questions. When classifying, the volunteers were presented with a randomly selected slice from the data volume (in conjunction with the two neighbouring slices above and below to create a ‘flipbook’ of five slices to provide some 3D context). The organelle of interest is then segmented by the volunteer in the online classification interface, using their mouse or stylus. Once an image has been annotated by a certain number of volunteers, it is removed from circulation and no longer passed to volunteers for segmentation (Fig. 2).
One of the core principles of citizen science is ‘the wisdom of the crowd’. That is, while an individual volunteer may be less accurate than an expert (although we definitely have some super-users who are every bit as good as the experts!), multiple volunteer annotations on the same image will, when aggregated together to form a consensus, produce answers of extremely high quality. These collective segmentations can then be used either directly, e.g. for addressing a specific research question, or as ground truth for training deep-learning systems, since this produces a larger quantity of ground truth data than is generally attainable via conventional means. Our model, trained with exclusively volunteer data, is as good as (or, arguably, even better) than those based on the usual expert annotation approaches, thanks to our ability to obtain larger quantities of training data.
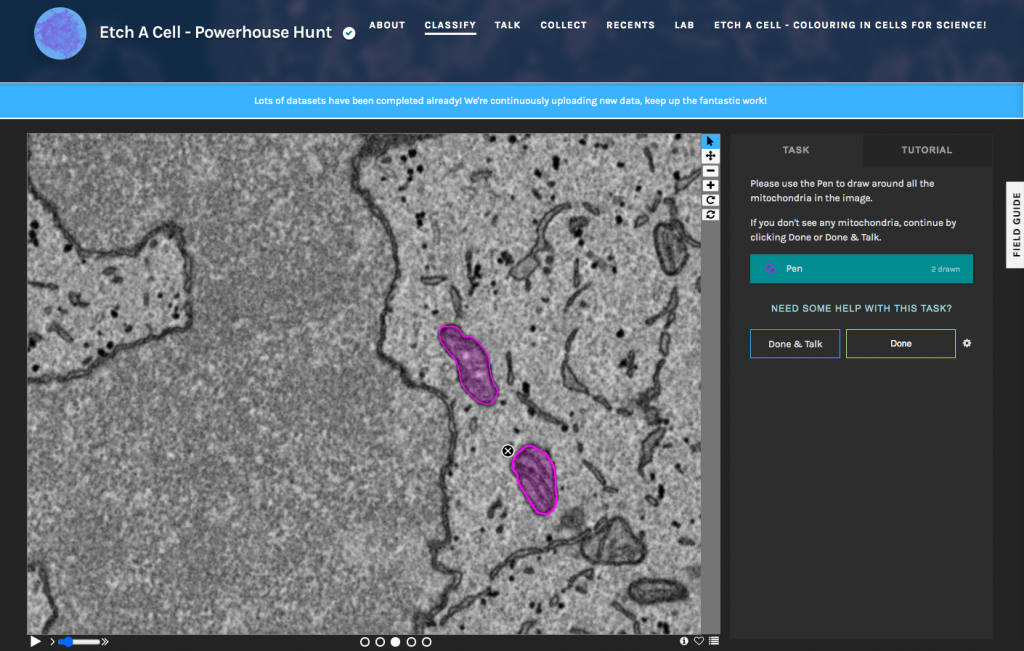
A key goal of our work is to be able to generalise the deep-learning models to work across a range of samples, microscopes, and imaging conditions. Whereas an expert can easily identify the same structures across multiple different datasets, automated methods frequently struggle due to subtle differences in the image properties. To achieve this generalisability, training data must be gathered for each variation so that the computer can learn which features are important, like the organelle to be segmented, and which should be ignored, like noise and artifacts. Again, the luxury of expert segmentations across a broad range of conditions is extremely rare; however, citizen science provides us with a promising avenue to generating these vast data sets. With this in mind, we are already developing additional Etch A Cell projects to tackle other organelles and conditions, including a project to segment mitochondria (Etch A Cell – Powerhouse Hunt). All our current projects and workflows can be found through our Etch a Cell organization page, and there are several more projects in the pipeline, so watch this space!
Beyond enabling novel data analysis, one of the most rewarding aspects of running an online citizen science project has been the opportunity to directly connect with the volunteers of all ages and backgrounds from around the world. Citizen science provides a valuable mechanism to engage many people with authentic science, giving people an insight into the day-to-day work of scientists dealing with raw data, rather than just the polished end-product they might be used to seeing in the usual press-releases and news stories.
For anyone considering building a citizen science project, the Zooniverse has a set of comprehensive guides to how to set up and run a project at https://help.zooniverse.org/. For further information, you can get in touch with the Zooniverse team at contact@zooniverse.org (Fig. 3).


Helen Spiers
Biomedical Research Lead of Zooniverse
The Francis Crick Institute, London, UK
Department of Astrophysics, University of Oxford, UK

Martin Jones
Deputy Head of Microscopy Prototyping
Electron Microscopy Science Technology Platform (EM STP)
The Francis Crick Institute, London, UK


 (2 votes, average: 1.00 out of 1)
(2 votes, average: 1.00 out of 1)