SRRF-Stream+ Super-Resolution Microscopy Accessible to All
Sponsored by Andor, on 12 August 2020
Fast, reliable & live-cell compatible Super-Resolution
Science has limits imposed by the laws of physics that constrain discoveries and the advance of knowledge. In microscopy, up until the beginning of the XXI century, the diffraction limit of light was an unbreakable barrier. This law of physics imposes that two points could not be resolved (clearly separated) if they are closer together than half the wavelength of light used to view them. In practice, this would mean that light microscopes could only resolve objects/structures that are separated by 200 nm or more. The 200 nm barrier would therefore leave a significant gap of knowledge to be uncovered, since many subcellular structures and organelles are smaller than 200 nm.
In the 1st decade of the XXI century, the diffraction limit of light was overcome using a number of ingenious microscopy techniques. A new field of discovery was ready to be revealed. Several methods for super-resolution evolved, allowing imaging beyond the diffraction limit of light. Super-resolution microscopy methods such as STED (stimulated emission depletion microscopy), STORM/PALM (Stochastic optical reconstruction microscopy)/ (Fluorescence photo-activation localisation microscopy) and SIM (Structured illumination microscopy) have become available for researchers.
Although the Super-resolution methods have opened a new field of discovery, some limitations on these methods existed. Complex sample preparation, long acquisition times as well as high energy requirements for acquisition, render these techniques inappropriate (if not incompatible) for use with live-cell imaging. Furthermore, the optical requirements and computer power needed render these SR methods extremely expensive and inaccessible to many labs.
More recent advances with spinning disk confocal technology combining optical over-sampling with computational reassignment do achieve a 1.6x improvement beyond the diffraction limit and are suitable for live-cell imaging, but at the significant cost of a reduced field of view, and starting from a lower native resolution. Additionally, it is still inextricably linked to the high-cost hardware.
The Dragonfly spinning disk confocal with its unique combination of pinhole size and custom pinhole pitch separation has an improved native axial and lateral resolution, delivering better resolution for routine imaging. Then, using photon reassignment (deconvolution) on the acquired images, the final obtained axial and lateral resolution of the dragonfly can be 240 nm and 139 nm respectively.
But what if a researcher needs more resolution? Or does not have a spinning disk confocal microscope? Is there an alternative method that is fast, compatible with live imaging, and that can still acquire super-resolved images at the full field of view and, if required, deep inside the cells? The answer to these questions is yes, with SRRF or SRRF-Stream+ (Figure 1).
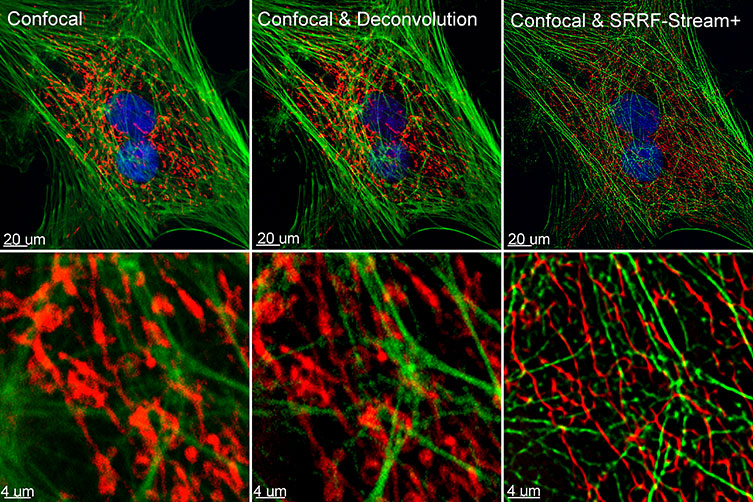
In 2016, the Henriques lab developed an alternative approach to super resolution named SRRF – Super-Resolution Radial Fluctuations [1]. SRRF can be combined with widefield, TIRF or confocal, and the final resolution will depend on the proprieties of the acquired data sets. Using the SRRF algorithm, the researchers can achieve resolutions in XY up to 50 nm [1]. Furthermore, the SRRF algorithm does not require special sample preparation or special fluorophores for acquisition, being compatible with conventional fluorophores and fluorescent proteins.
Most importantly, a super-resolved SRRF image can be acquired by capturing on average between 20-100 frames (more frames will result in improved resolution) and the energy required for this imaging is in the order of the mW to W per cm2 range, making SRRF compatible with live-cell imaging. [1,2] SRRF was initially provided as an ImageJ plugin. In order to obtain a SRRF image, a long acquisition workflow needed to take place and the SRRF-image was obtained by post-acquisition processing of the acquired data in the image J plugin – NanoJ.
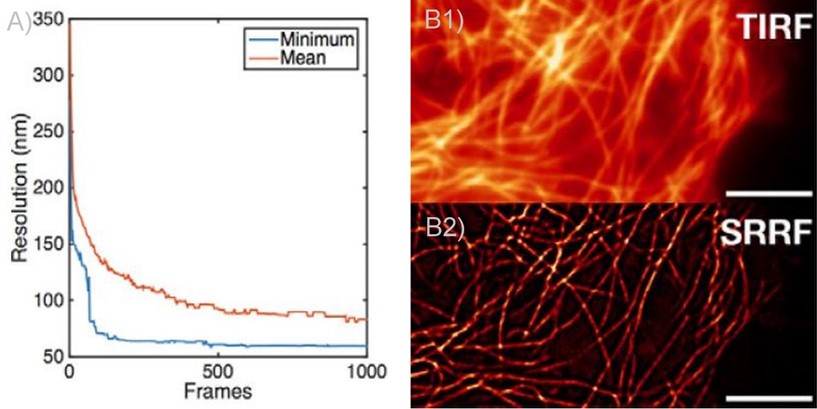
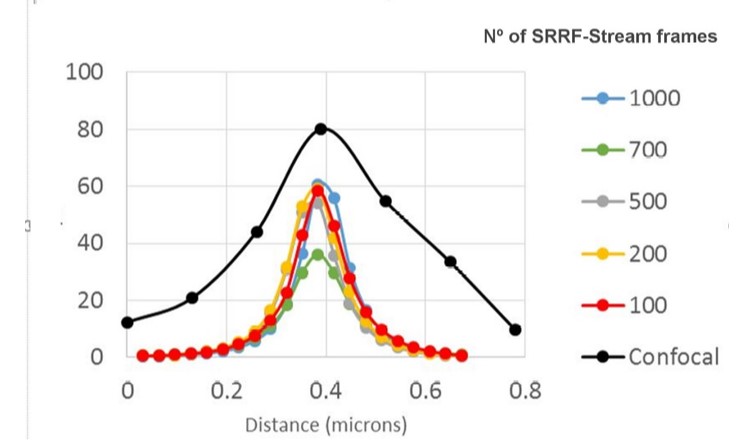
In 2018 from collaboration between Andor and professor Henriques, Andor launched SRRF-Stream. SRRF-Stream is Andor´s implementation of the SRRF algorithm that delivers super-resolution images on the fly with the click of a button. As with the original SRFF algorithm, the resolution of SRRF-Stream will improve with the number of frames captured per a given time point. This is most striking up to 100 frames, reaching a more stable increase from 100 frames to 500 frames, and a very modest improvement in resolution from 500 frames onwards (figures 2, 3). Please note that other factors will also affect resolution such as exposure time, Nyquist sampling, radiality magnification and the ring radius increase; these should be tested in order to optimise results.
ANDOR´s SRRF-Stream was a major improvement allowing live-cell super-resolution, super-resolution deep inside cells or tissues way beyond the top of the coverslip.
SRRF-Stream is available exclusively with iXon-EMCCD cameras, through micro-manager and Fusion which is the control software for Dragonfly confocal. The advantages of SRRF-Stream are [3,4]:
- Allows Real-Time super-resolution images, enhancing the workflow and avoids post-processing.
- Breaks the diffraction limit of light by delivering Super-Resolved images with 2 to 6-fold increase in the final resolution (50-150 nm), depending on the imaging mode used (i.e. widefield, confocal or TIRF) and the experimental conditions.
- Use of Low Excitation power Intensities for imaging (mW to W/cm2), which results in higher compatibility with extended live cell observations and with minimal impact in cell physiology.
- Compatible with conventional Fluorophore and fluorescent proteins.
- Easy sample preparation.
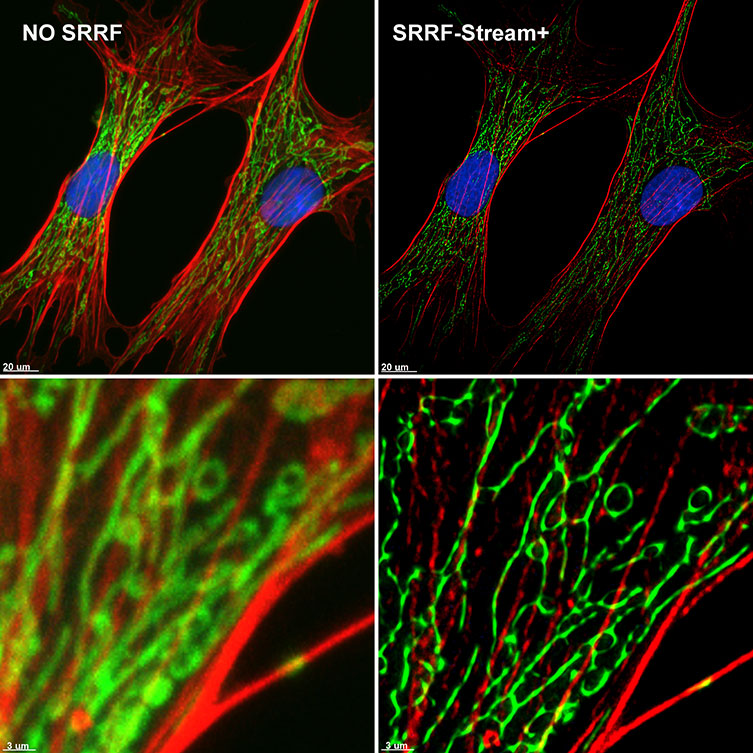
SRRF-stream is, therefore, a cost-effective solution for any microscope since all of them can be converted into super-resolution microscopes. SRRF-stream is compatible with any imaging modality, namely widefield, TIRF and confocal (Figure 4).
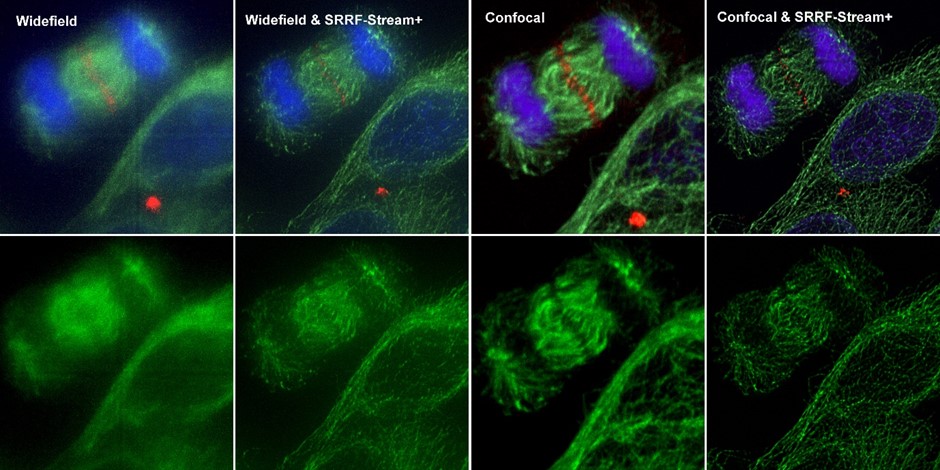
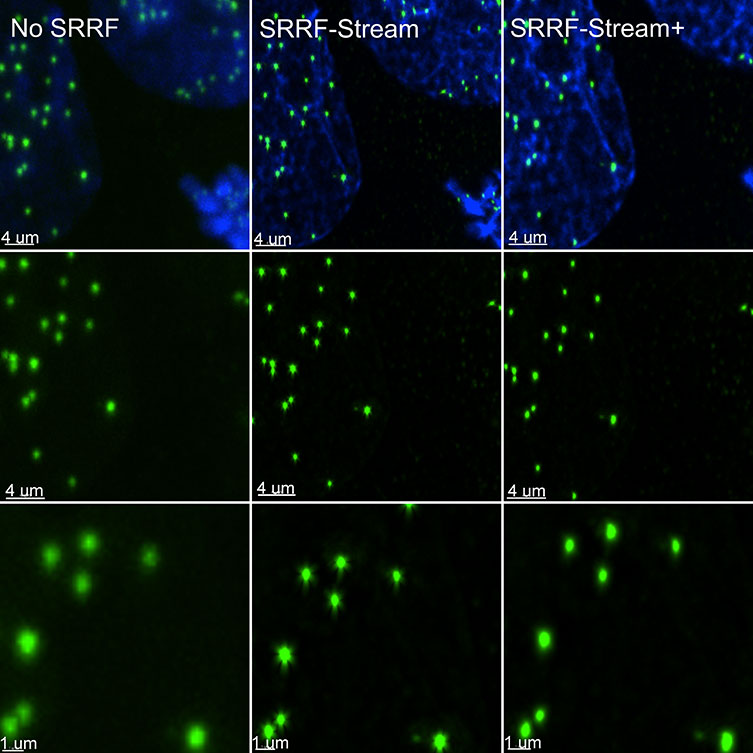
This July 2020 we are proud to announce an additional improvement to our SRRF-Stream algorithm: SRRF-Stream+ (figures 1, 4, 5, 6, 7).
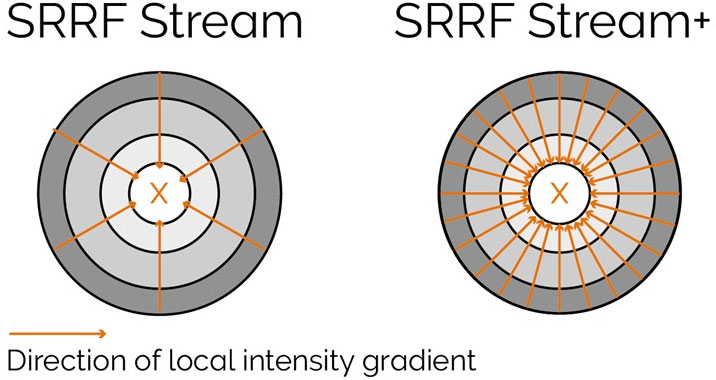
SRRF-Stream+ goes a step further with increased radiality measurements. On the previous version of SRRF, the radiality measurements were computed in 6 directions and now are computed in 24 directions (Figure 5). This enhancement will improve the result of your super-resolved processing data, and the occasional star artefacts previously visible, especially in circular structures such as kinetochores, are now removed (Figure 7).
To get the best from SRRF-Stream+, as before, the user will need to acquire images under Nyquist criterium. As previously reported, the major benefit in resolution will be obtained when oversampling by a factor of 2.3 or higher. Still, when oversampling by a factor of 1.5 good results can yet be obtained using the SRRF-Stream+ (for more detailed information on how to sample on Ixon cameras please go to Andor original SRRF-stream tech note).
When analysing Figures 1, 4, 5 and 7 the benefit on SRRF-Stream+ is obvious, but one question will come into the mind of many potential users: How is the speed of acquisition affected? If SRRF-stream+ computes radialities in 24 direction instead of 6; is SRRF-Stream+ 4X slower than the original version of SRRF? When developing the new SRRF algorithm, Andor took into account that speed of acquisition is also an essential parameter to many researchers. It has been possible to achieve the improved image quality with a minimal impact to the acquisition speeds. To achieve this, CUDA performance has been optimised, perfectly utilising the massive computing power of nVidia graphic card to speed up SRRF-Stream+ calculations. As a result, a time-lapse of 100 time points with 50 SRRF-stream+ images per time point (corresponding to 500 images in total) will only vary by 0.5 secs in acquisition when comparing with our original SRRF-stream (full field of view (1024 X1024) images acquired with Ixon Ultra).
As for Andor original SRRF-Stream advantages, they are maintained in SRRF-Stream+ but with the improved image quality. Super-resolution imaging processing is still carried out at ultra-fast processing rates (up to 30X faster) than ImageJ SRRF (“NanoJ-SRRF’), and SRRF image acquisition/processing is captured on the fly, in parallel to the acquisition of the data. SRRF-Stream+ algorithm delivers images with a final resolution between 50-150 nm and due to its low energy requirements is an ideal solution for live-cell microscopy. Super resolved images can be acquired using with large fields of view, and importantly super-resolution microscopy is not restricted to cell surface events. With SRRF-Stream+, it´s possible to acquire a super-resolved image deep inside cell or tissues, and there is no need for special sample preparation.
Since it’s launch, SRRF-Stream has become widely used for many applications. Examples of using SRRF-Stream in iXon cameras include: to analyse mitochondria trafficking into cell terminus via microtubule destabilisation [5], or analysis of HIF1α nuclear translocation in mesenchymal stem cells [6]. The Dragonfly multimodal confocal system was also be used to acquire SRRF-Stream images, examples are: analysis of cytokinesis using optogenetic tools [7] as well as vesicle trafficking studies on analysing the effects of class II PI3Ks in the regulation of clathrin-dependent pinocytosis [8]. Other applications of SRRF-Stream+ include analysis of protein structure at a sub-organelle level, tracking of single molecules inside cells, membrane fusion studies of individual SNARE protein machinery, intracellular skeleton reassembly (changes to actin fibre meshwork).
In conclusion, we have improved our SRRF-Stream algorithm delivering live-cell compatible super-resolution with even better results than before. SRRF-Stream+ delivers super-resolved images of intracellular structures without artefacts, can compensate for the effects of cameras fixed pattern noise, and delivers high-quality super-resolved images.
Many other applications and discoveries are waiting to be revealed using SRRF-Stream+. Do you want to know more about SRRF-Steam+? Contact us we will be delighted to show it in action.



 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)

