Asymmetric cell divisions in 3 dimension stem cell colonies
Posted by Agathe Chaigne, on 27 September 2021
When looking at the development of a multicellular organism, for example a human, the first striking feature is the progressive increase in cell numbers due to successive divisions, from one single tiny cell 80 mm in diameter to a 3.5 kg ball of organised cells forming a new-born baby. The way cells divide, in particular the orientation and position of the division plane, which define the respective positions and sizes of the two nascent daughter cells, can be crucial to determine daughter cell fate. For instance, in early mouse development division orientation determines in part which cells become part of the embryo itself, and which become part of the extra-embryonic tissues like the placenta (Zhang and Hiiragi, 2018).
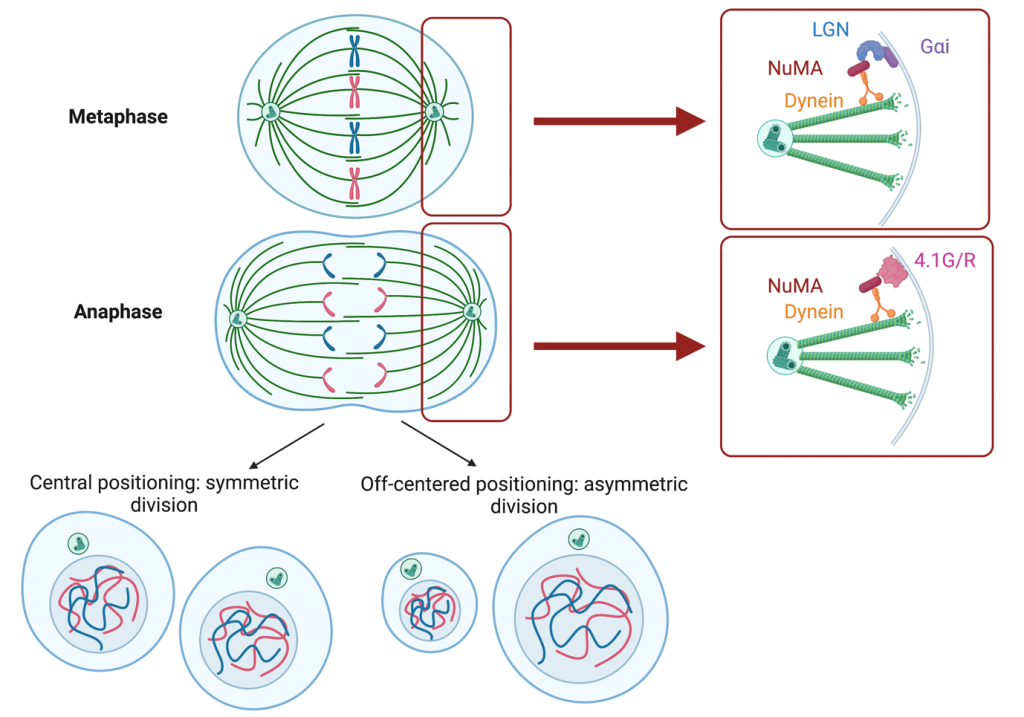
What we know about the control of the orientation and positioning of the division plane in mammalian cells comes, for the most part, from the study of single cancer cells and flat epithelial tissues. In these 2-dimension contexts, we know that the position of the division plane is determined by the position of the mitotic microtubule spindle in metaphase (Figure 1). Astral microtubules emanating from the spindle contact the cortex on both sides. The cortex can be decorated with two versions of a protein complex containing the spindle positioning protein NuMA: NuMA/LGN/GaI (in metaphase) and NuMA/4.1G/R (in anaphase) (Kiyomitsu and Cheeseman, 2013; McNally, 2013). In both these complexes, NuMA interacts with motors that walk along astral microtubules, pushing or pulling the spindle and positioning it within the cell. If the spindle is positioned at the centre of the cell, the division will be symmetrical, if it is off-centred, the two daughter cells will have different sizes (Figure 1). In many cell types, the localization of the NuMA containing protein complexes depends on the shape of the cell in interphase, and this results in the spindle being positioned along the cell’s interphase long axis (Bosveld et al., 2016; Fink et al., 2011; Théry et al., 2005; Théry et al., 2007). In contrast to this detailed understanding of spindle positioning in 2D contexts, we do not know much about the mechanisms for spindle positioning and daughter cell size control during cell division in 3D contexts. Yet, division in 3D is essential during early mammalian development, when the embryo is essentially a ball of cells, and in many adult tissues in both physiological and pathological contexts.
How to image cell division in 3D?
When I started my postdoc in Ewa Paluch’s lab at the MRC LMCB, University College London, we decided to investigate this question using mouse embryonic stem cells as a model system, because they grow in 3D colonies (Figure 2) and undergo fate transitions that resemble what happens in a mammalian embryo (Kalkan et al., 2017). The first challenge was to be able to follow cell division in these 3D colonies. Live cell imaging always involves minimizing phototoxicity, which is possible by finding a good balance between having a good time resolution and spatial resolution and not imaging the cells with too much laser power or too often.
Furthermore, I needed to be able to find dividing cells to image. Usually, people studying cell division maximize the number of cells they can analyse by synchronizing the cells (for example by pre-treating the cells with a CDK1 inhibitor- when this inhibitor is washed out, the cells all undergo mitosis). In 3D colonies of mouse embryonic stem cells, synchronising cells lead to disruption of the structure of the colony due to synchronous rounding of too many mitotic cells, which appeared to destabilise colony structure. Thus, to ensure sufficient numbers of dividing cells without perturbation, I decided to image multiple colonies- to the great sadness of the IT personnel that had to cope with terabytes of data. Ultimately, with help from Andrew Vaughan at the imaging facility of the LMCB, we settled for multi-position acquisition of Z-stacks (with 1 um between planes) of cell colonies, every 5 minutes using a spinning-disc microscope.
How to segment cells in 3D?
The second challenge that we came across was to be able to accurately measure cell size and quantify spindle position in 3D- there are many tools allowing measurement of 2D parameters, such as area, or shape, but this is a much more complicated problem when looking at a 3D blob of cells. Fortunately, I could team up with Matthew Smith, another postdoc in Ewa’s lab at the time, who developed a plugin to segment dividing cells in 3D (Smith et al., 2017). This plugin, freely available for Fiji (Schindelin et al., 2012), is based on a closed triangulated meshwork that can deform to minimise the image energy, which its does, for example, by deforming towards the regions of high intensity .
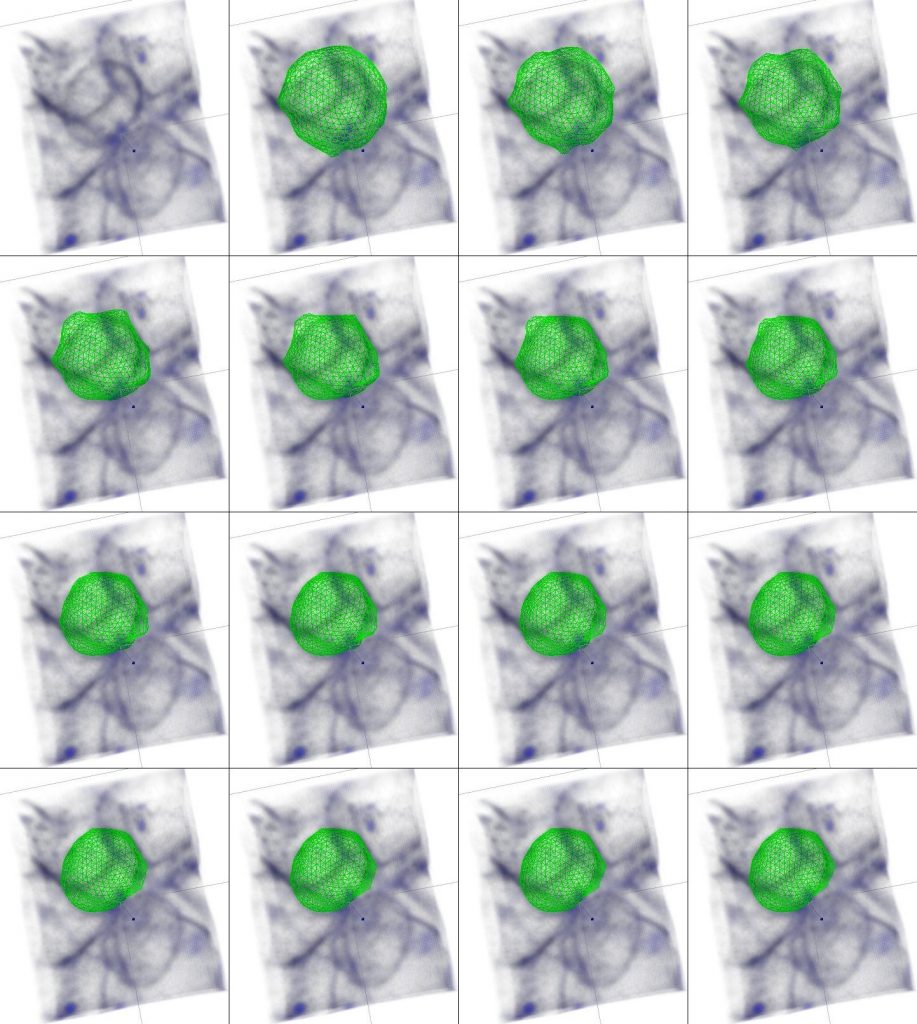
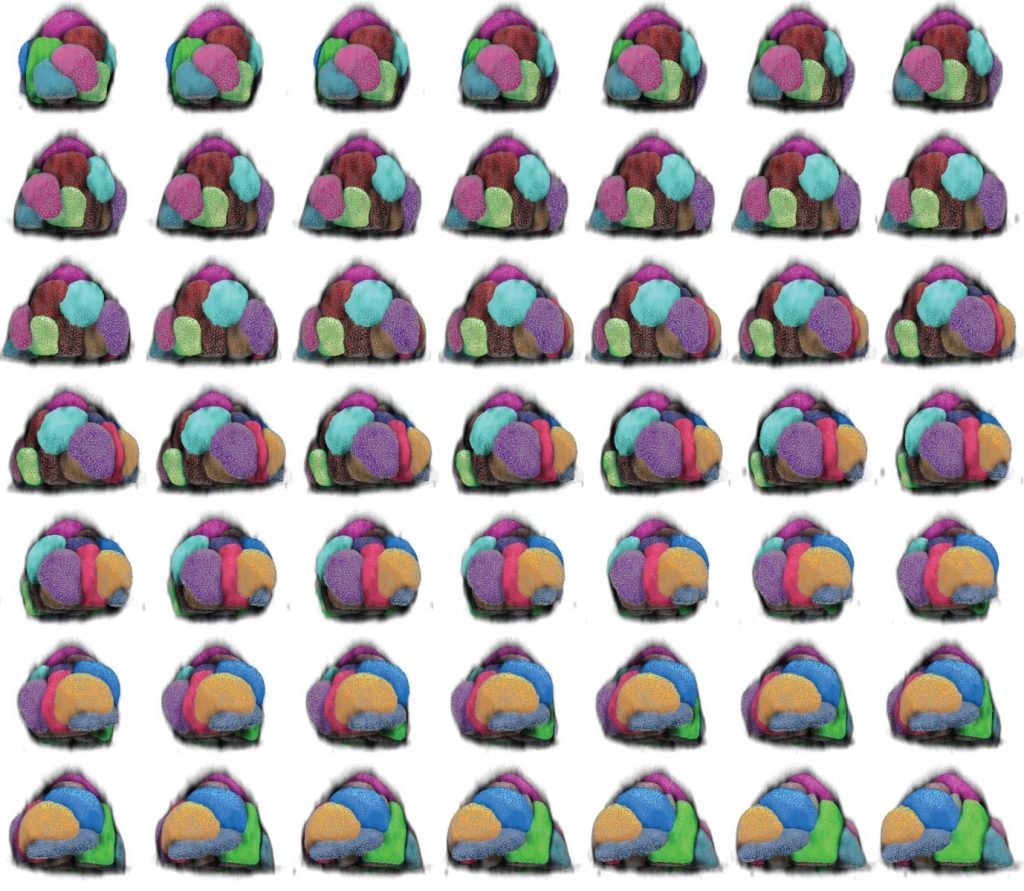
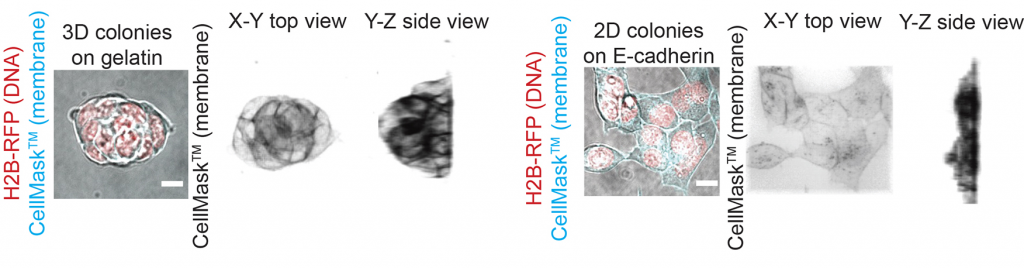
It is thus possible, for example using a membrane dye, to get the mesh to closely surround the cell contour and thus obtain a 3D segmentation of a cell (Figure 2) or a whole colony of cells (Figure 3).
Naïve embryonic stem cells divide asymmetrically
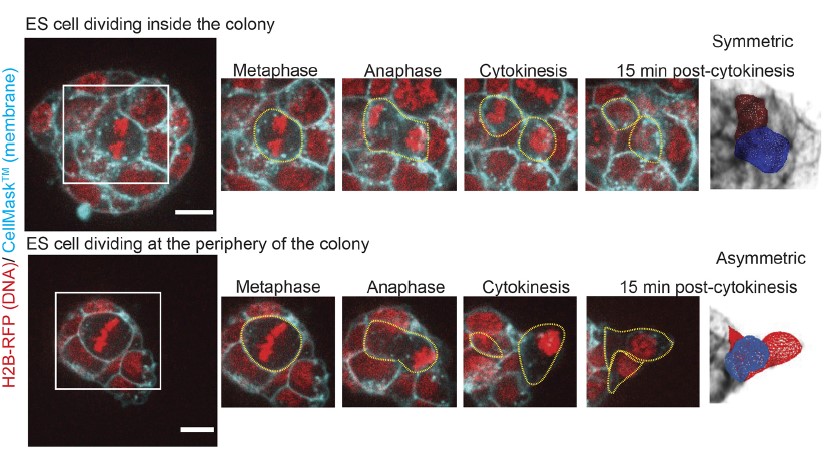
Equipped with this set of techniques, we explored spindle positioning and daughter cell size control during stem cell division in 3D colonies (Chaigne et al., 2021). We realised that while cells divided symmetrically when they were inside the colony, cells that divided at the periphery of the colony divided asymmetrically in size, with one daughter cell being bigger than the other (Figure 4). What could explain the difference between cells dividing inside and at the periphery? As mentioned earlier, spindle positioning is a major driver of daughter cell size control during mitosis; we thus decided to investigate spindle positioning. We used our 3D segmentation plugin to record the position and orientation or the metaphase plate within the cell as a proxy for spindle position. We discovered that the spindle moves more in all directions in cells dividing peripherally, and is more stable in cells dividing inside the colonies. This prompted us to ask what was the difference between cells inside and at the periphery of the colony.
Cell-cell junctions are an important player in the control of daughter cell size after division
A major difference between cells dividing inside and at the periphery of a colony is the distribution of the cell-cell adhesion protein E-Cadherin. Indeed, inside a colony, a dividing cell has a homogeneous distribution of E-Cadherin at its surface. In contrast, cells dividing at the periphery of a colony are in contact with other cells only on part of their surface, which often results with one of the nascent daughter cells being firmly in contact with the colony while the other at least partially not (Figure 4). We reasoned that plating the cells directly on E-Cadherin would allow both daughter cells to have similar contact with E-Cadherin through their bottom surface, as all cells would be spread on a flat E-Cadherin substrate (Figure 5). We observed that this was sufficient to both reduce spindle motion in cells dividing at the periphery of these flat colonies, and to reduce the proportion of asymmetrically dividing cells, which suggested to us that indeed, the amount of E-Cadherin at the cell surface during division controls spindle mobility and daughter cell size. At this point, we do not know, however, how E-Cadherin could control spindle stability!
What happens to division symmetry during early differentiation?
In the embryo, embryonic stem cells undergo a fate transition whereby they exit from naïve pluripotency and become receptive to differentiation signals before committing towards a specific lineage (Kalkan et al., 2017). In culture, when embryonic stem cells exit pluripotency, they also undergo a drastic shape change, from the 3D colonies we studied so far to a 2D sheet of spread cells (De Belly et al., 2021). We noticed that while cells exited pluripotency, division became more symmetric, with the two daughter cells being approximatively the same size. We wondered what the mechanism was for this transition between asymmetric and symmetric division, and we turned to the obvious pathway for spindle positioning, involving the cortical complexes containing NuMA (Figure 1). We found that interestingly, NuMA was not recruited at the cortex in metaphase stem cells or at any point of exit from pluripotency. However, NuMA was progressively recruited to the cortex in anaphase when cells exited pluripotency, suggesting that it could help stabilise the spindle at the centre of the cells in anaphase, similarly to what has been observed in HeLa cells (Figure 1, (Kiyomitsu and Cheeseman, 2013)).
Stem cells divide along their metaphase long axis
Stabilisation of the spindle in the middle of HeLa cells depends on NuMA recruitment at the cortex, which also aligns the division plane with the interphase long axis (Figure 1). We thus wondered whether the orientation of cell division in stem cells also correlated with cell shape. To investigate this, we turned back to our 3D segmentation plugin, that also allows us to quantify features of cell shape such as elongation. We noticed that naïve stem cells in 3D colonies did not have very elongated shapes at any point of their cell cycle and did not align their division plane with their long axis. However, when cells exited pluripotency, they became more elongated, including during cell division; this correlated with an alignment of the spindle with the metaphase long axis, rather than the interphase long axis. This suggests that in stem cells, spindle positioning might not be controlled by interphase shape but that maybe metaphase shape could instruct NuMA recruitment in anaphase.
Conclusion
Based on our observations, we propose a model where a cross-talk between cell-cell junctions and mitotic cell shape controls spindle positioning in stem cells. Naïve cells in 3D colonies have very mobile spindles, possibly because of heterogeneities in E-Cadherin junctions distribution between nascent daughter cells. Cells remain very spherical throughout division and (maybe as a consequence) do not recruit NuMA in anaphase, and divide asymmetrically. When cells exit naïve pluripotency, they drastically change shape and spread. They adopt elongated shapes, recruit NuMA in anaphase, and divide symmetrically. An obvious- but still hypothetical- model could be that during exit from naïve pluripotency, the decrease in cell-cell junction strength stabilises the spindle at the centre of the cells, and the shape elongation we observed in metaphase directs recruitment of NuMA at the cortex in anaphase, further stabilising the spindle and rendering division more symmetrical. Specific mechanisms that link E-Cadherin to spindle motion, and NuMA recruitment to cell shape are still unknown. One of the key questions remaining is the role of these asymmetric divisions. I have shown that differences in size between sister cells have no consequence in the dynamics of pluripotency exit (Chaigne et al., 2020). Our most striking finding is possibly how little control naïve stem cells appear to have over their division. It could be that having a loose control of division, and as a result a high level of division asymmetries, introduces noise in the system in terms of cell cycle stage and cell size. This could in turn be important for cell fate transitions at the population level, as increase in noise and heterogeneity has been shown to be important for the dynamics of fate transitions in tissue remodelling (Petridou et al., 2021) and specification (Romanos et al., 2020). How division asymmetries in embryonic stem cells affect lineage commitment during differentiation remains to be investigated.
References
Bosveld, F., Markova, O., Guirao, B., Martin, C., Wang, Z., Pierre, A., Balakireva, M., Gaugue, I., Ainslie, A., Christophorou, N., et al. (2016). Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature 530, 495–498.
Cadart, C., Zlotek-Zlotkiewicz, E., Le Berre, M., Piel, M. and Matthews, H. K. (2014). Exploring the Function of Cell Shape and Size during Mitosis. Developmental Cell 29, 159–169.
Chaigne, A., Labouesse, C., White, I. J., Agnew, M., Hannezo, E., Chalut, K. J. and Paluch, E. K. (2020). Abscission Couples Cell Division to Embryonic Stem Cell Fate. Developmental Cell.
Chaigne, A., Smith, M. B., Lopez Cavestany, R., Hannezo, E., Chalut, K. J. and Paluch, E. K. (2021). Three-dimensional geometry controls division symmetry in stem cell colonies. Journal of Cell Science 134, jcs255018.
De Belly, H., Stubb, A., Yanagida, A., Labouesse, C., Jones, P. H., Paluch, E. K. and Chalut, K. J. (2021). Membrane Tension Gates ERK-Mediated Regulation of Pluripotent Cell Fate. Cell Stem Cell 28, 273-284.e6.
Fink, J., Carpi, N., Betz, T., Bétard, A., Chebah, M., Azioune, A., Bornens, M., Sykes, C., Fetler, L., Cuvelier, D., et al. (2011). External forces control mitotic spindle positioning. Nature Cell Biology 13, 771–778.
Kalkan, T., Olova, N., Roode, M., Mulas, C., Lee, H. J., Nett, I., Marks, H., Walker, R., Stunnenberg, H. G., Lilley, K. S., et al. (2017). Tracking the embryonic stem cell transition from ground state pluripotency. Development 144, 1221–1234.
Kiyomitsu, T. and Cheeseman, I. M. (2013). Cortical Dynein and Asymmetric Membrane Elongation Coordinately Position the Spindle in Anaphase. Cell 154, 391–402.
McNally, F. J. (2013). Mechanisms of spindle positioning. J. Cell Biol. 200, 131–140.
Petridou, N. I., Corominas-Murtra, B., Heisenberg, C.-P. and Hannezo, E. (2021). Rigidity percolation uncovers a structural basis for embryonic tissue phase transitions. Cell S0092867421001677.
Pinheiro, D., Hannezo, E., Herszterg, S., Bosveld, F., Gaugue, I., Balakireva, M., Wang, Z., Cristo, I., Rigaud, S. U., Markova, O., et al. (2017). Transmission of cytokinesis forces via E-cadherin dilution and actomyosin flows. Nature 545, 103–107.
Romanos, M., Allio, G., Combres, L., Médevielle, F., Escalas, N., Soula, C., Steventon, B., Trescases, A. and Bénazéraf, B. (2020). Cell-to-cell heterogeneity in Sox2 and Brachyury expression ratios guides progenitor destiny by controlling their motility. Developmental Biology.
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682.
Smith, M. B., Chaigne, A. and Paluch, E. K. (2017). An active contour ImageJ plugin to monitor daughter cell size in 3D during cytokinesis. In Methods in Cell Biology, pp. 323–340. Elsevier.
Théry, M., Racine, V., Pépin, A., Piel, M., Chen, Y., Sibarita, J.-B. and Bornens, M. (2005). The extracellular matrix guides the orientation of the cell division axis. Nature Cell Biology 7, 947–953.
Théry, M., Jiménez-Dalmaroni, A., Racine, V., Bornens, M. and Jülicher, F. (2007). Experimental and theoretical study of mitotic spindle orientation. Nature 447, 493–496.
Zhang, H. T. and Hiiragi, T. (2018). Symmetry Breaking in the Mammalian Embryo. Annual Review of Cell and Developmental Biology 34, 405–426.


 (No Ratings Yet)
(No Ratings Yet)