Imaging nuclear dynamics and chromatin organization in a live intact organism: the design of the minimal constraint device
Posted by Dana Lorber, on 9 March 2022
To visualize the response of the nucleus and chromatin to mechanical signals, I developed a device that enables their imaging in a live intact Drosophila larva. Together with genetically encoded fluorescent markers, we can track muscle contraction and nuclear dynamics, as shown in Movie 1. While the muscle is at rest, we can image chromatin in great detail, as shown in movie 2.
The minimal constraint device facilitates the acquisition of spatiotemporal data from a live intact organism. In the supplementary material of the quoted papers [1, 2, 3], you will find additional examples that show microtubule deformation, Ca2+ entry to the muscle cytoplasm, and nuclear lamina deformation, as well as details on chromatin organization.
In this post, I will take you through the device design process and some of the problems encountered during the development of the minimal constraint device.
The development of the minimal constraint device
The device was designed for imaging contractile muscles and follow concomitant nuclei dynamics, in a live intact Drosophila larva, with minimal interference to muscle physiology and dynamics and with no external stimulation.
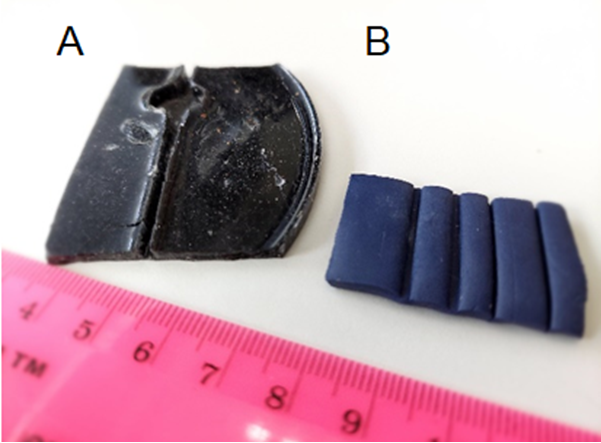
I realized that to image the larva during natural movement, its body muscles should be able to contract without body crawling. This was achieved by attaching both larval ends to thin glass capillaries, by placing the tip of a capillary in a drop of superglue (which fills its end by capillary action), and its attachment to the larva, with a very small amount of glue (the capillary’s inner diameter is 198 μm). Improving the adhesion process requires temporary numbing of the larva (with CO2 or by cooling) to allow the alignment of the two capillaries with the larval body during attachment. I used polymer clay from a hobby shop and Sylgard casts to create molds with grooves of various widths to find the proper conditions for the alignment (Figure 1). During attachment, the larva is placed in the groove, and the capillaries slide in the groove towards the larval edges until they touch and adhere to the larva (Figure 2).

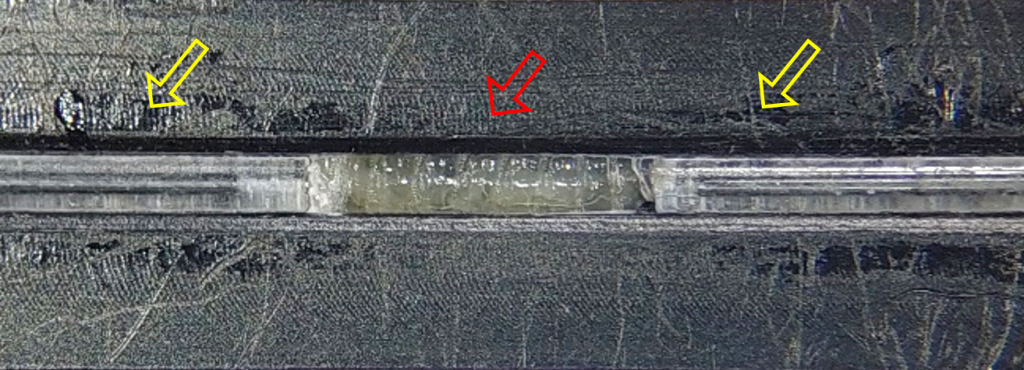
Next, we designed and manufactured a machine-made device (made by the manufacturing unit at the Weizmann Institute of Science), based on the same principles, to enable precise fit of the device to the microscope stage, especially in terms of working distance from the lens.
To track myonuclei during muscle contraction, imaging took place when the larva was in the groove, preventing it from bending sideways but still allowing it to contract freely. During muscle contraction and shortening, the nuclei changed position in the Z direction and sometimes went off the microscope focal plane. To minimize this effect, I filled the gaps between the groove and the larva with hydrogel, reducing out-of-plane events while maintaining larval free contractions. I chose to use alginate, a hydrogel produced from brown seaweed from the North Sea that polymerizes promptly in the presence of divalent molecules at room temperature. The mechanical properties of the gel can be tuned by changing the amount of dry material and the choice of cross-linking cations [4].
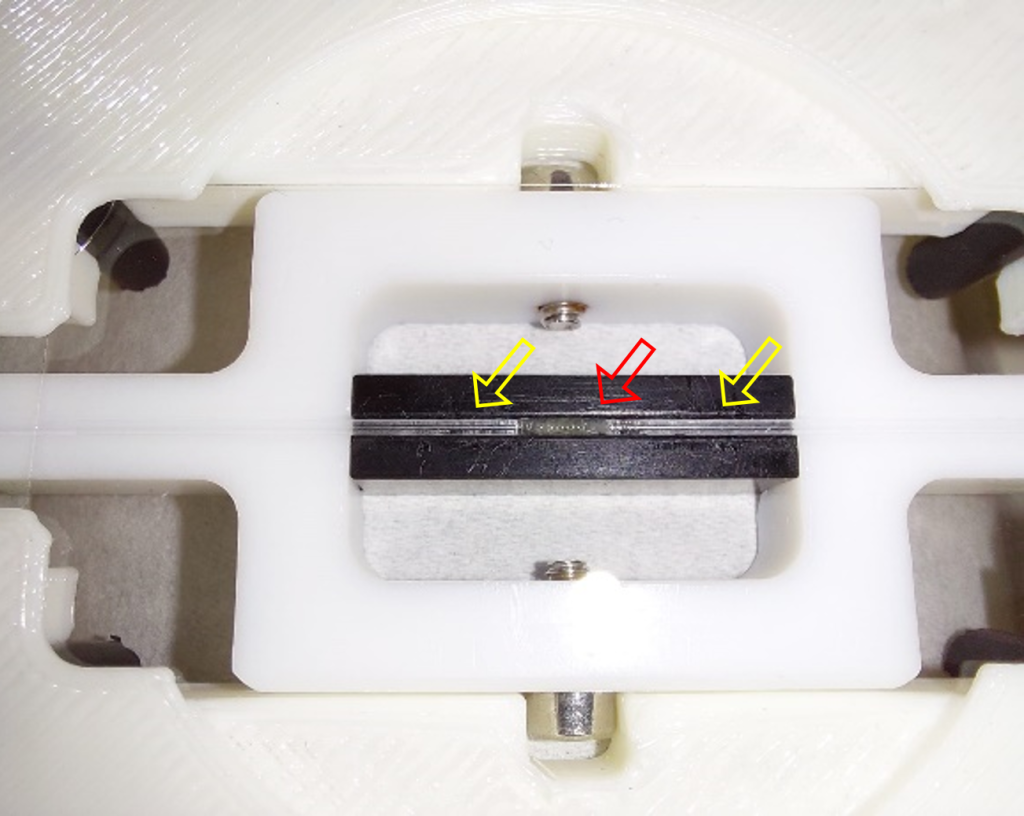
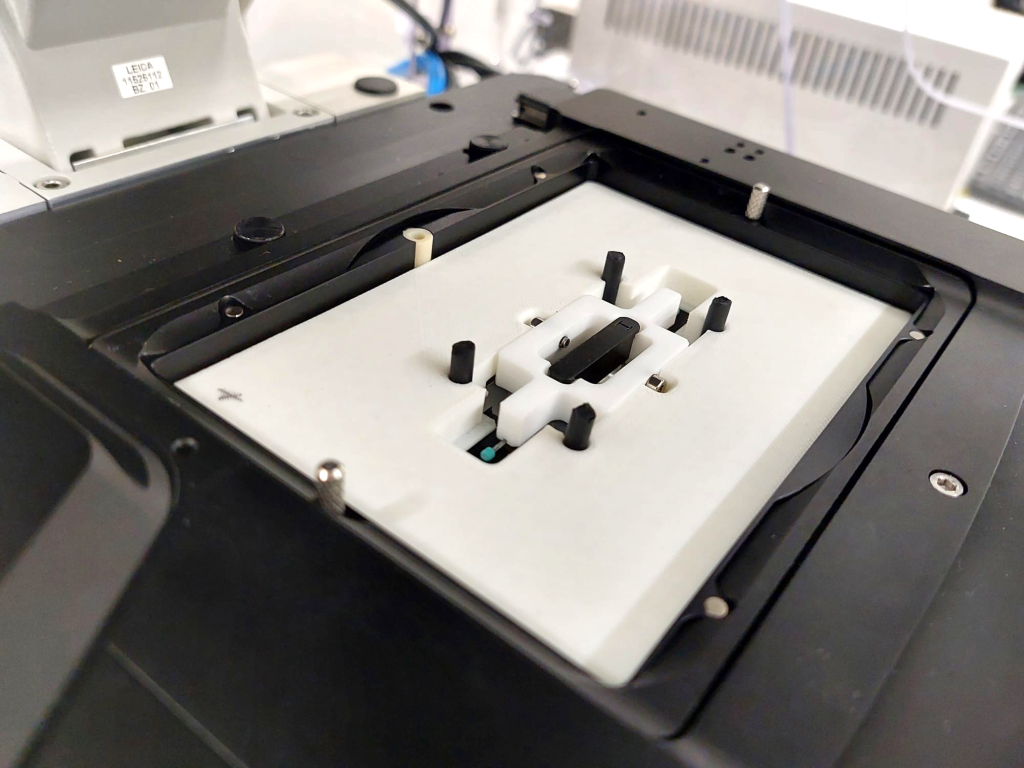
The final design of the minimal constraint device is detailed in Figure 3. The larva is awake throughout the experiment with minimal interference with its movement, allowing to image a whole muscle and its nuclei during muscle contraction. When the muscles are stationary, nuclei and chromatin can be imaged in their native tissue.
Using the device to study myonuclei in the live intact model
In our first study using this device, we explored the dynamics of muscle nuclei during muscle contraction of a mutant larva lacking a functional “Linker of Nucleoskeleton and Cytoskeleton (LINC)” complex (Nesprin/klar mutant), that connects muscle nuclei to the sarcomeric cytoskeleton [1]. We found that in this mutation, the mechanical dynamics of the myonuclei changed and lost their uniformity along the muscle fiber, possibly preventing nuclei synchronized activity found in the control muscle. In the following work, using this device, we studied 3D chromatin organization under physiological conditions and found that chromatin organizes at the nuclear periphery, leaving a chromatin-devoid volume at the center of the nucleus, contrary to the prevailing model [2]. Furthermore, we demonstrated no radial separation between active and repressed chromatin as they reside side-by-side at the nuclear periphery. Interestingly, peripheral chromatin organization was also observed in freshly isolated human neutrophils and cultured effector T cells, suggesting that peripheral chromatin organization is not Drosophila-specific but is evolutionarily conserved.
In conclusion, the minimal constraint device enables visualization and quantification of nuclear position and structure dynamics during muscle contractile/relaxation waves, as well as alterations in chromatin organization.
Dana Lorber
Research Associate
Lab of Talila Volk
Dept Molecular Genetics
Weizmann Institute of Science
References
1. Dana Lorber, Ron Rotkopf, and Talila Volk, Lab on a Chip 20, 2100 (2020). https://doi.org/10.1039/D0LC00214C
2. Daria Amiad-Pavlov*, Dana Lorber*, Gaurav Bajpai, Adriana Reuveny, Francesco Roncato, Ronen Alon, Samuel Safran, and Talila Volk, Science Advances 7, (2021). 10.1126/sciadv.abf6251
3. Dana Lorber and Talila Volk, APL Bioengineering. 010902, (2022). https://doi.org/10.1063/5.0069286
4. Jose Guisan, editor, Immobilization of Enzymes and Cells, second edition (Humana Press Inc., 2006).
*authors contributed equally


 (No Ratings Yet)
(No Ratings Yet)