FocalPlane playlist: Christophe Leterrier
Posted by Christophe Leterrier, on 14 November 2023
Playlist part 1: recent articles and preprints
The first part of my playlist is made of articles and preprints that I found particularly interesting during the last months. As bioRxiv is nearing its 10th anniversary, we can all agree how transformative preprints have been for biology in general, and for the thriving bioimaging community at the heart of the FocalPlane audience. One of the most useful series on FocalPlane for me is the regular recap of microscopy-related preprints (https://focalplane.biologists.com/tag/preprints/) – despite following the literature quite closely, there is always a couple of interesting works that I had missed in there!
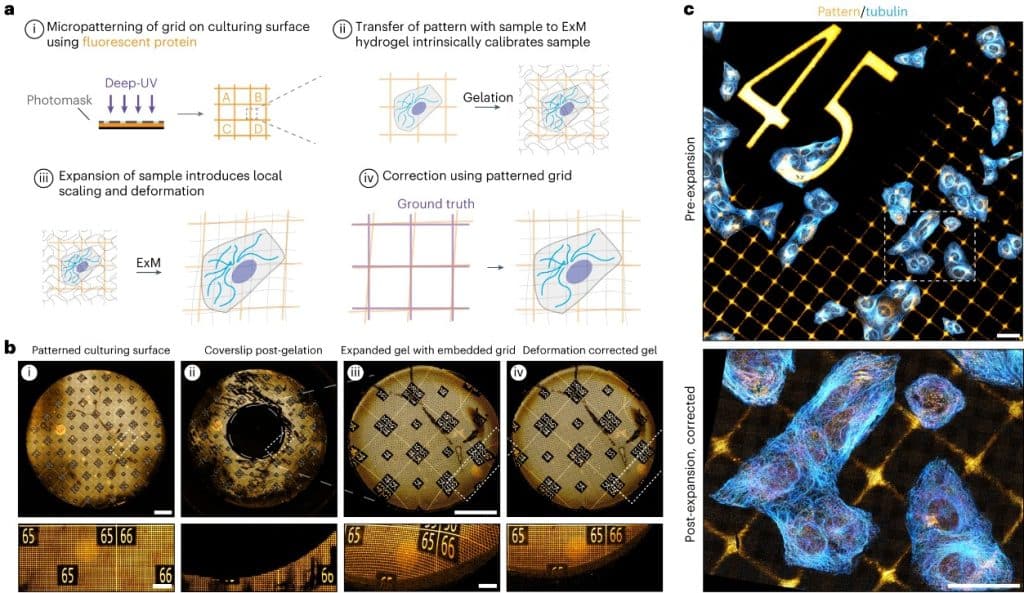
GelMap: intrinsic calibration and deformation mapping for expansion microscopy
Damstra HGJ, Passmore JB, Serweta AK, Koutlas I, Burute M, Meye FJ, Akhmanova A, Kapitein LC.
Nat Methods. 2023;20(10):1573–80, previously preprinted on bioRxiv
Expansion microscopy is arguably the most significant innovation in microscopy in the last 10 years. I like that it was primarily an innovation in sample preparation rather than optical hardware. Its performance and ease of use has driven adoption in hundreds of labs that could access the nanoscale detail of cellular organization using regular fluorescence microscopes. It is still one of the most exciting techniques to follow these days, with lots of innovation to refine the techniques and broaden their applicability. In this article, Damstra et al., from the lab of Lukas Kapitein in Utrecht, have developed a technique to print registration grids that are preserved after expansion of cell cultures and brain slices, allowing for fine registration of the expanded sample, and alleviating the common concern about local deformations during the expansion procedure.
Rho GTPase signaling and mDia facilitate endocytosis via presynaptic actin
Oevel K, Hohensee S, Kumar A, Rosas-Brugada I, Bartolini F, Soykan T, Haucke V.
bioRxiv. 2023 Oct 16.
Another example of how progress in labeling is often crucial to open new possibilities in microscopy is the tagging of endogenous proteins using CRISPR-Cas9. In this recent preprint, Oevel et al., from the lab of Volker Haucke in Berlin, have carefully examined the role of actin in endocytosis at presynapses. Among the strategies used, they have specifically labeled endogenous presynaptic actin using small tags and imaged it using multicolor STED. We have previously used a similar endogenous actin tagging strategy in our work delineating the distinct actin nanostructures within presynapses (Bingham et al. J Cell Biol 2023, previously on bioRxiv).
Correlative single-molecule and structured illumination microscopy of fast dynamics at the plasma membrane
Winkelmann H, Richter CP, Eising J, Piehler J, Kurre R.
bioRxiv. 2023 Oct 9.
Our lab has primarily used Single Molecule Localization Microscopy (SMLM) since its creation in 2017, imaging the nanoscale organization of the neuronal cytoskeleton using STORM and PAINT on fixed cells. These days I am really interested in adding a dynamic aspect to our work, and Structured Illumination Microscopy (SIM) is really the best widely-available compromise to obtain super-resolved images on fragile living cells. There is a lot of ongoing development on the processing side, with classical and deep learning-based reconstruction algorithm allowing to minimize the amount of excitation light – another field where the acceleration provided by preprints and open source development are decisive. In this recent preprint, Winkelmann et al., from the Piehler and Kurre labs in Osnabrück, have managed to optimize SIM acquisition speed to combine it with single-molecule tracking, obtaining magnificent data on the diffusion of receptors at the plasma membrane relatively to the organization of the cortical actin network.
Super-Resolution Imaging of Fast Morphological Dynamics of Neurons in Behaving Animals
Zhang Y, Bai L, Wang X, Zhao Y, Zhang T, Ye L, Du X, Zhang Z, Du J, Wang K.
bioRxiv. 2023 Oct 24.
SIM works great on thin samples like cultured cells on glass, but it can also be pushed to obtain super-resolved data on more thick samples, and even in living animals! In this recent work from the Wang lab in Shanghai, Zhang et al. have used an orthogonal light-sheet geometry combined with structured excitation to image the detailed morphology of dendritic spines and axonal boutons in brain slices and living mouse brains.
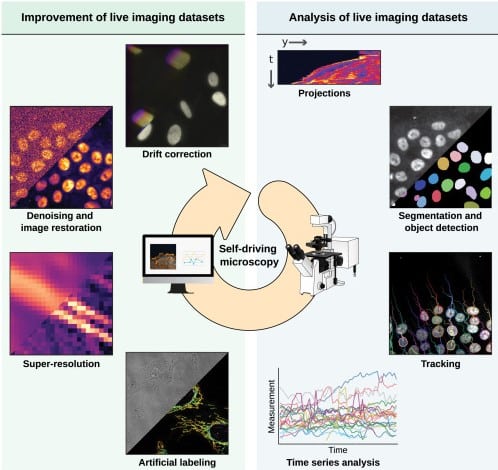
Live-cell imaging in the deep learning era
Pylvänäinen JW, Gómez-de-Mariscal E, Henriques R, Jacquemet G.
Curr Opin Cell Biol. 2023;85:102271.
Another hot topic these days is the use of deep learning for image processing and analysis, which emerged in the last 5 years as a strategy to bypass the compromises between spatio-temporal resolution and photodamage in live-cell imaging. This recent review from the labs of Guillaume Jacquemet in Turku and Ricardo Henriques in Lisbon (with whom I’ve had the pleasure to collaborate on a couple of image analysis tools, our latest here, preprinted here) is a great summary of the current methods and their uses, highlighting how “computational microscopy” is quickly becoming one of key technique in our field.
De novo protein identification in mammalian sperm using in situ cryoelectron tomography and AlphaFold2 docking
Chen Z, Shiozaki M, Haas KM, Skinner WM, Zhao S, Guo C, Polacco BJ, Yu Z, Krogan NJ, Lishko PV, Kaake RM, Vale RD, Agard DA.
Cell. 2023;(186):1–13, previously preprinted on bioRxiv
Finally, I want to finish this article playlist by shifting away from optical microscopy and mention the incredible cryo-EM works that pop up these days. What is happening right now is a revolution, fueled by progress in three-dimensional electron microscopy of native-state protein complexes in their cellular context. Adding on top of that, the way to assign the identity of the protein components, thanks to a “reverse AlphaFold” approach, resolves the issue with protein tagging, and allows reaching a true “structural biology in cells” approach. I’m very excited to see what will come next, particularly in neurons!
Playlist part 2: great FocalPlane content
In this second part of the playlist, I’d like to highlight content from FocalPlane which I think demonstrate its uniqueness for today’s microscopy-inclined biologists. FocalPlane posts often fill a unique need by providing topical, appealing information on our community and its tools.
Latin American Microscopists interviews (by Mariana de Niz)
I really like the “Latin American Microscopists” blog series made by Mariana De Niz. She interviews microscopists from each Latin America country as well as Latin American scientists working all over the world. I learn a lot not only about the science, but also about the variety of paths taken by these researchers, the challenges and the joys of doing science. Kudos to Mariana for this series which will soon reach its 100th entry!

60 years of fluorescent proteins (by Joachim Goedhart)
This short post by Joachim Goedhart not only provides a timeline for the development of fluorescent proteins from the initial jellyfish GFP to the latest variants such as mStayGold, but also explains how he used the programming language R to generate this timeline. Joachim is a great educator and tool developer – I’m a fan of his posts on FocalPlane and its sister site the Node, as well as his statistics webapp (available at https://huygens.science.uva.nl/)
A schedule for organizing international symposia (by Robert Haase)
This blog post exemplifies the kind of knowledge that a website such as FocalPlane can bring – more substantial than tweets, less formal than academic article: what are the different steps in organizing a symposium, and when to plan them? This is a post I wish I had read when I helped organize my first meeting a couple of years ago. Its author Robert Haase is a pillar of the bioimage analysis community, and I encourage you to also check his more technically-inclined posts about important and sometimes overlooked aspects such as licenses, data hosting and user documentation (https://focalplane.biologists.com/author/robert-haase/).
Have you ever dreamt of wandering around in a cell, walking in microtubules, hitchhiking on a mitochondrion? This is now possible thanks to Beata Mierzwa, who has been working at the interface of science, art and communication. Learn more about the Microscopya videogame on her blog post, and try it for free on your computer (https://beatascienceart.itch.io/microscopya) or on your phone!



 (No Ratings Yet)
(No Ratings Yet)