DL4MicEverywhere – Overcoming reproducibility challenges in deep learning microscopy imaging
Posted by Iván Hidalgo-Cenalmor, on 29 July 2024
Deep learning easy, accessible, and reproducible for everyone, everywhere
Recently, we released DL4MicEverywhere1 to enable biologists to apply their AI workflows for bioimage data analysis in reproducible research and in a user-friendly manner across different systems. DL4MicEverywhere’s containerisation of deep learning approaches facilitates their use across computers so the methods become fully reproducible and reusable in the long term. DL4MicEverywhere is easy to use and gives access to an extensive deep learning platform with state-of-the-art models for a variety of image processing tasks. You can read more about our new baby in Hidalgo-Cenalmor, I. et al., Nat Methods 2024 and start using it on its website: DL4MicEverywhere GitHub.
👇Curious about how this works and why it matters? You’re in the right place!👇
The Quest for the Reproducible DL Pipeline: A short story
Once Upon a Time, …
… an intrepid researcher was on a mission to create a bioimage analysis pipeline in Python.
After countless hours of effort and perseverance, the code finally ran like a charm.
Thrilled, the researcher shared its code on GitHub for the world to access, report bugs, and contribute to its evolution. However, joy was short-lived: users reported missing packages and version conflicts. Ah, yes, the pipeline required additional Python packages!
Promptly, the researcher uploaded a requirements.txt file listing the necessary packages and versions, the dependencies.
The Curse of Changing Dependencies
Months passed, and disaster struck.
The researcher ventured into new workflows, only to discover that the once-reliable pipeline had stopped working. An error message taunted: “The function you want to run does not exist for your package”.
But fear not! The researcher discovered conda environments, isolated Python setups that manage project dependencies separately, ensuring that each project installation remains unaffected by the others. An environment.yaml file was added to GitHub, enabling others to replicate the appropriate conda environment.
The Compatibility Conundrum
A new issue was opened on GitHub: “I tried installing your pipeline, but conda tells me that the packages are not available in the conda channel arm64. I’m using an M1 Mac. Thank you”.
The researcher, bound by the limitations of their Windows machine, could only offer a helpless apology, unable to replicate the error.
As if the compatibility challenges with different operating systems (OS) weren’t enough, another issue was received: “Thank you for this amazing contribution. I’ve been using it for a while, but I got a new laptop, and when I try installing it again, it tells me that some packages are no longer available.”
What steps could our researcher take to address these new challenges?
The Containerised Solution
The researcher decided on one last attempt: virtual machines.
Virtual machines emulate separate computational environments enabling isolated installations; like a virtual computer inside your main computer. A specific type of virtual machine is a container, such as those created by Docker2, which emulate these “independent boxes”, each with its own computational properties. Docker uses Docker images as protocols to build containers. From a single Docker image, countless containers can be spawned, each sharing the exact same computational environment, including Python version installations. This ensures that a project runs consistently, no matter where, guaranteeing long-term reproducibility3.
A containerisation approach for their current and future pipelines was born: “DL4MicEverywhere”, a foundation for reproducibility in the field of deep learning and microscopy.
The story behind DL4MicEverywhere
DL4MicEverywhere builds upon the previous work, ZeroCostDL4Mic4, which has enabled the image data analysis of a variety of research studies5,6,7, gained significant popularity in recent years among life scientists and trainers8,9, and inspired new developments across the community10,11,12. The main idea behind ZeroCostDL4Mic was to use Jupyter notebooks that followed a consistent fully documented structure, where the code is hidden, requiring the user to provide only simple input information (e.g., data paths or patch size for training). This approach allowed biologists with limited programming skills to utilize advanced deep learning methods. However, each of the notebooks relies on numerous libraries and dependencies that are constantly updated with new features. We developed DL4MicEverywhere to address these challenges and extend the applicability of ZeroCostDL4Mic by making user-friendly pipelines reproducible everywhere, regardless of the computer or installation.
What’s new with DL4MicEverywhere?
To ensure the long-term reproducibility and cross-compatibility of deep learning workflows across computers, we use Docker containers to encapsulate each notebook along with its dependencies. This setup allows running notebooks outside Google Colab, by using Jupyter Lab.
To achieve this, we developed an automated program that checks each notebook and its dependencies, then installs and packages them into a Docker image. This Docker image is then used to launch the Docker container on the user’s machine. Once created, the Docker image provides a static environment where dependencies and algorithms are fixed. Moreover, DL4MicEverywhere gives access to the entire ZeroCostDL4Mic platform of deep learning approaches by making its notebooks reproducible across systems.
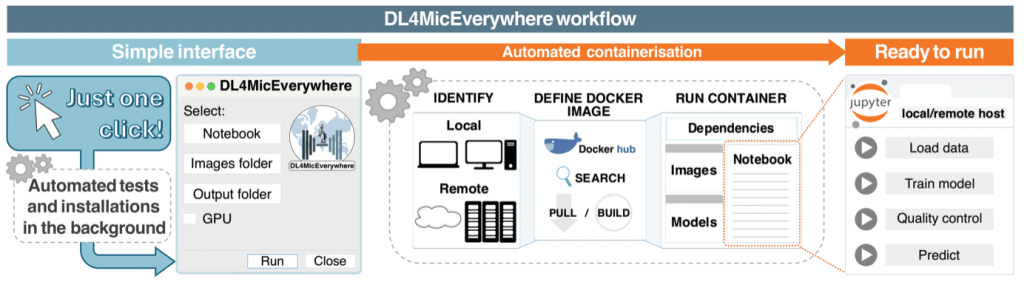
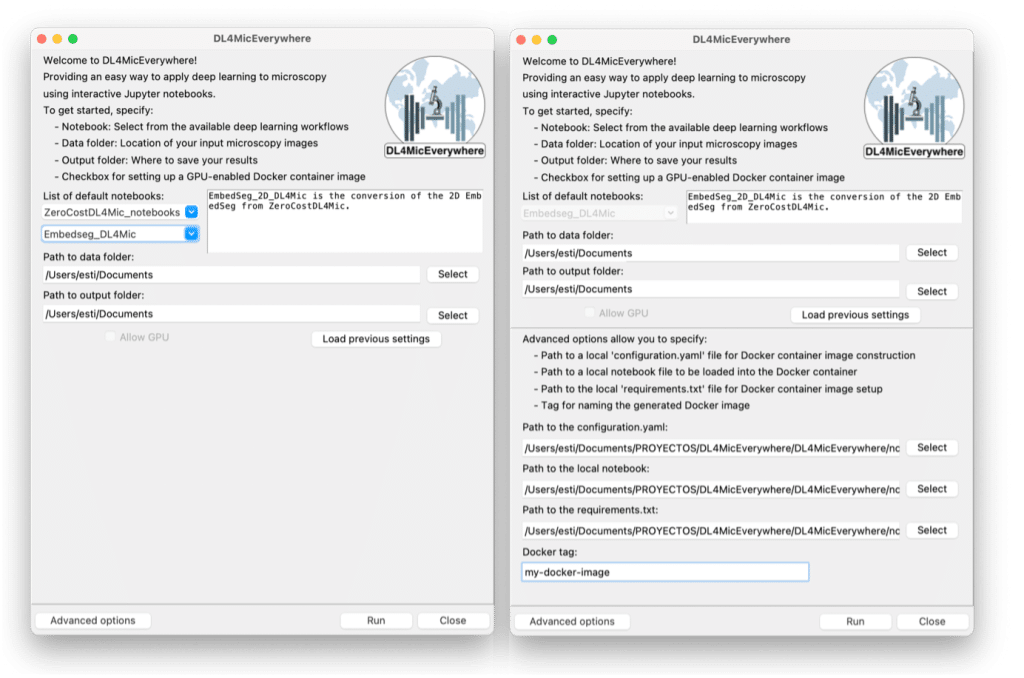
This encapsulation and installation workflow (see Figure 2) is hidden behind a user-friendly GUI that guides the user through selecting the notebook and specifying the paths to the data to analyze. The GUI has a basic mode (see Figure 3a) for existing configurations and an advanced mode (see Figure 3b) for custom setups.
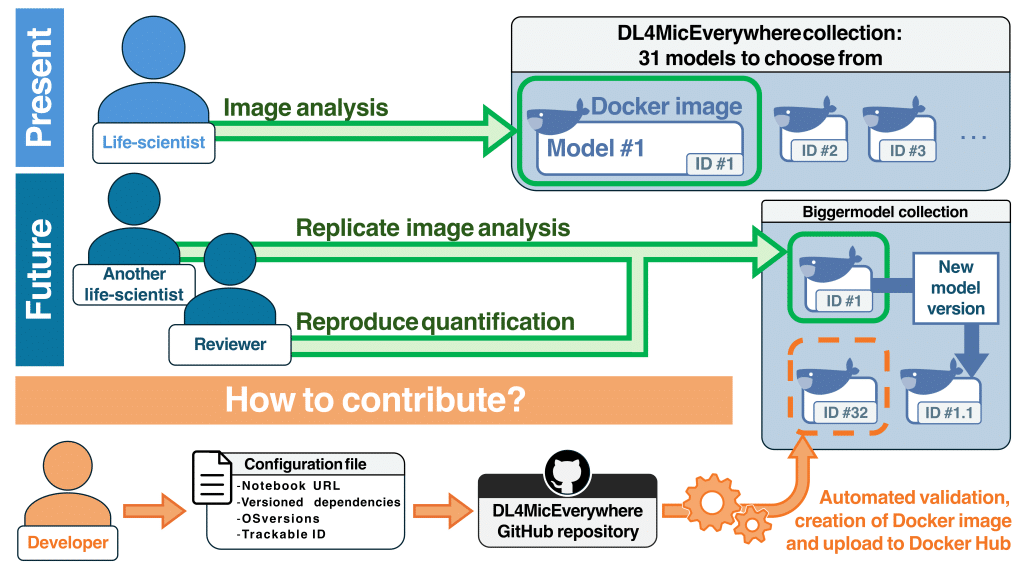
We defined containerisation standards that enabled becoming a community partner of BioImage Model Zoo13 and maintaining a collection of trustable notebooks together with versioned, validated, and published Docker images. These images are available on Docker Hub and are linked to the notebooks using a unique identifier that supports version traceability.
Why should DL4MicEverywhere matter to you?
It’s all about long-term reproducibility and replicability of analytical results without dealing with programming geekiness. Here are a few scenarios (see Figure 4) where this can be useful for you:
- Research continuity: DL4MicEverywhere ensures that your analytical pipelines remain up-to-date and functional, even after a year or more, by allowing you to easily find and use the exact version you originally used. This continuity is crucial for research continuity and ensures that your methods remain reliable and reproducible.
- Easy review process and reproducibility of published results: DL4MicEverywhere simplifies the review process by providing a unique identifier for the deep learning approach used. This ensures that reviewers can replicate the analytical pipeline, improving the reviewing process and ensuring that after publication, the results remain reproducible.
- Enhancement of developers’ productivity: DL4MicEverywhere streamlines imaging workflows by automating processes and providing an easy contribution format. Developers can effortlessly integrate containerised methods in the DL4MicEverywhere collection, allowing the dissemination of their work to the community through the connection with the BioImage Model Zoo and the publication of validated and versioned Docker images. This allows you to focus on improving your methods, making your work reproducible and shareable with minimal efforts.
DL4MicEverywhere brings a new level of efficiency and reliability to deep learning in microscopy. It’s about making sure your methods stay useful and accessible, no matter how much time has passed or how much technology has changed.
How does DL4MicEverywhere work?
Dive into our documentation and video tutorials to gain a deeper understanding of its inner workings:
Step-by-step User Guide:
Explore our detailed document for a comprehensive walkthrough of DL4MicEverywhere’s functionality:
User Guide Video:
Watch this brief tutorial on YouTube to see DL4MicEverywhere in action.
Windows users:
If you’re using Windows, we’ve got you covered with dedicated tutorial videos tailored to your operating system.
Literature
1.- Hidalgo-Cenalmor, I., Pylvänäinen, J.W., G. Ferreira, M. et al. DL4MicEverywhere: deep learning for microscopy made flexible, shareable and reproducible. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02295-6
2.- Merkel, D. Docker: lightweight linux containers for consistent development and deployment. Linux j 239, 2 (2014).
3.- Moreau, D., Wiebels, K. & Boettiger, C. Containers for computational reproducibility. Nat Rev Methods Primers 3, 50 (2023). https://doi.org/10.1038/s43586-023-00236-9
4.- von Chamier, L. et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nature Communications 12, 2276 (2021). https://doi.org/10.1038/s41467-021-22518-0
5.- Matchett, K.P., Wilson-Kanamori, J.R., Portman, J.R. et al. Multimodal decoding of human liver regeneration. Nature 630, 158–165 (2024). https://doi.org/10.1038/s41586-024-07376-2
6.- Casey Eddington, Jessica K. Schwartz, Margaret A. Titus; filoVision – using deep learning and tip markers to automate filopodia analysis. J Cell Sci 15 February 2024; 137 (4): jcs261274. doi: https://doi.org/10.1242/jcs.261274
7.- Zelba, O., Wilderspin, S., Hubbard, A. et al. The adult plant resistance (APR) genes Yr18, Yr29 and Yr46 in spring wheat showed significant effect against important yellow rust races under North-West European field conditions. Euphytica 220, 107 (2024). https://doi.org/10.1007/s10681-024-03355-w
8.- NEUBIAS: https://eubias.org/NEUBIAS/training-schools/neubias-academy-home/, > 10 international courses and > 800 visitors per month to the platform resources
9.- Sivagurunathan, S., Marcotti, S., Nelson, C. J., Jones, M. L., Barry, D. J., Slater, T. J. A., Eliceiri, K. W., & Cimini, B. A. (2023). Bridging Imaging Users to Imaging Analysis – A community survey. Journal of Microscopy, 00, 1–15. https://doi.org/10.1111/jmi.13229
10.- Stringer, C., Wang, T., Michaelos, M. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106 (2021). https://doi.org/10.1038/s41592-020-01018-x
11.- Priessner, M., Gaboriau, D.C.A., Sheridan, A. et al. Content-aware frame interpolation (CAFI): deep learning-based temporal super-resolution for fast bioimaging. Nat Methods 21, 322–330 (2024). https://doi.org/10.1038/s41592-023-02138-w
12.- D. Franco-Barranco, J. A. Andrés-San Román, P. Gómez-Gálvez, L. M. Escudero, A. Muñoz-Barrutia and I. Arganda-Carreras, “BiaPy: a ready-to-use library for Bioimage Analysis Pipelines,” 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 2023, pp. 1-5. https://doi.org/10.1109/ISBI53787.2023.10230593
13.- Ouyang, W. et al. BioImage Model Zoo: A Community-Driven Resource for Accessible Deep Learning in BioImage Analysis. bioRxiv 2022.06.07.495102 (2022). https://doi.org/10.1101/2022.06.07.495102


 (No Ratings Yet)
(No Ratings Yet)
