Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 6 June 2023
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools.
Spatial redundancy transformer for self-supervised fluorescence image denoising
Xinyang Li, Xiaowan Hu, Xingye Chen, Jiaqi Fan, Zhifeng Zhao, Jiamin Wu, Haoqian Wang, Qionghai Dai
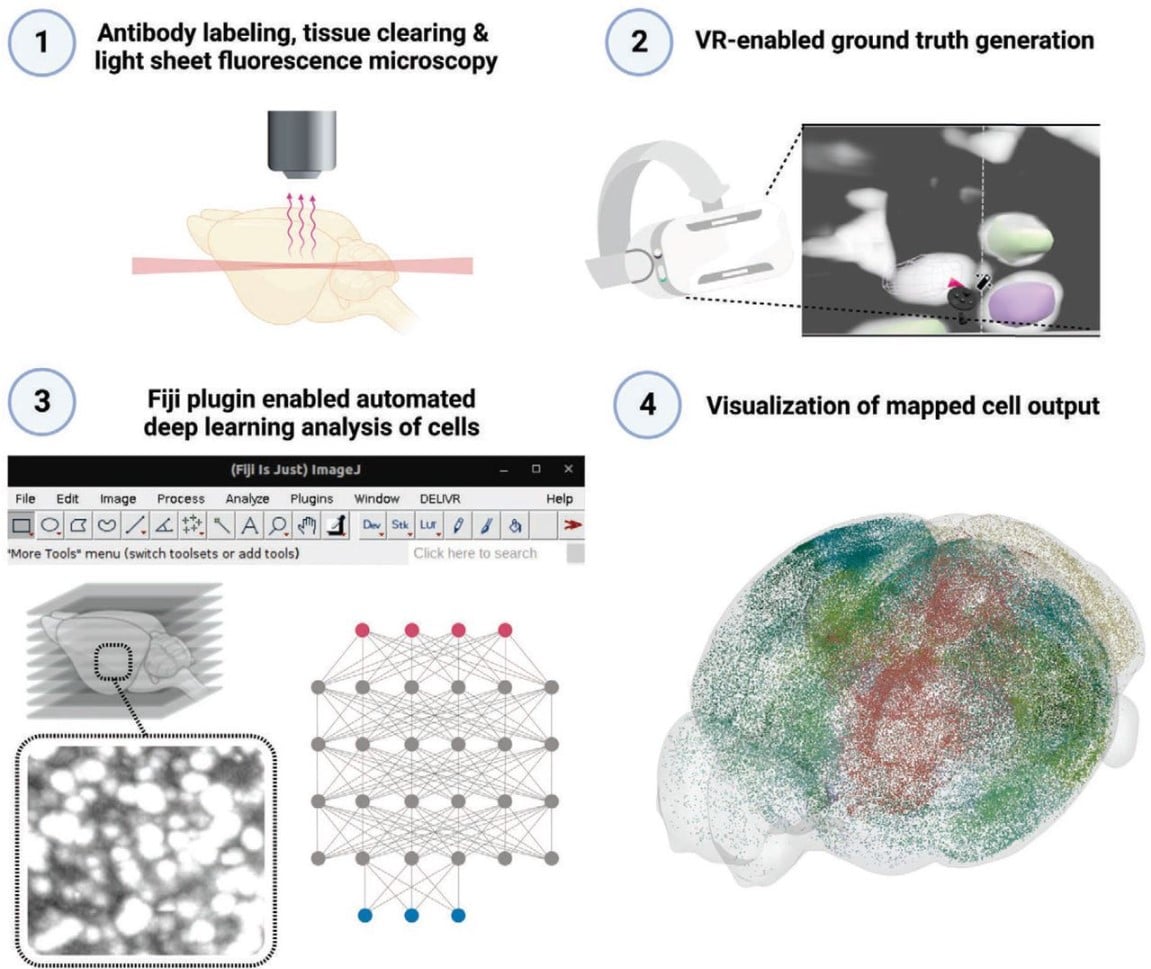
Virtual reality empowered deep learning analysis of brain activity
Doris Kaltenecker, Rami Al-Maskari, Moritz Negwer, Luciano Hoeher, Florian Kofler, Shan Zhao, Mihail Todorov, Johannes Christian Paetzold, Benedikt Wiestler, Julia Geppert, Pauline Morigny, Maria Rohm, Bjoern H. Menze, Stephan Herzig, Mauricio Berriel Diaz, Ali Ertürk
An image segmentation method based on the spatial correlation coefficient of Local Moran’s I
Csaba Dávid, Kristóf Giber, Katalin Kerti-Szigeti, Mihaly Kollo, Zoltán Nusser, László Acsády
But, what are the cells doing? Image Analysis pipeline to follow single cells in the zebrafish embryo
Arianne Bercowsky Rama, Olivier F. Venzin, Laurel Rohde, Nicolas Chiaruttini, Andrew C Oates
Self-supervised dense representation learning for live-cell microscopy with time arrow prediction
Benjamin Gallusser, Max Stieber, Martin Weigert
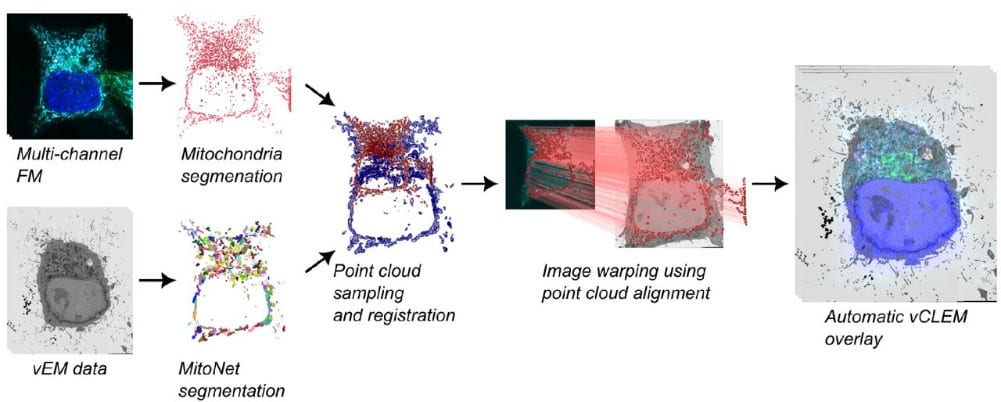
CLEM-Reg: An automated point cloud based registration algorithm for correlative light and volume electron microscopy
Daniel Krentzel, Matouš Elphick, Marie-Charlotte Domart, Christopher J. Peddie, Romain F. Laine, Ricardo Henriques, Lucy M. Collinson, Martin L. Jones
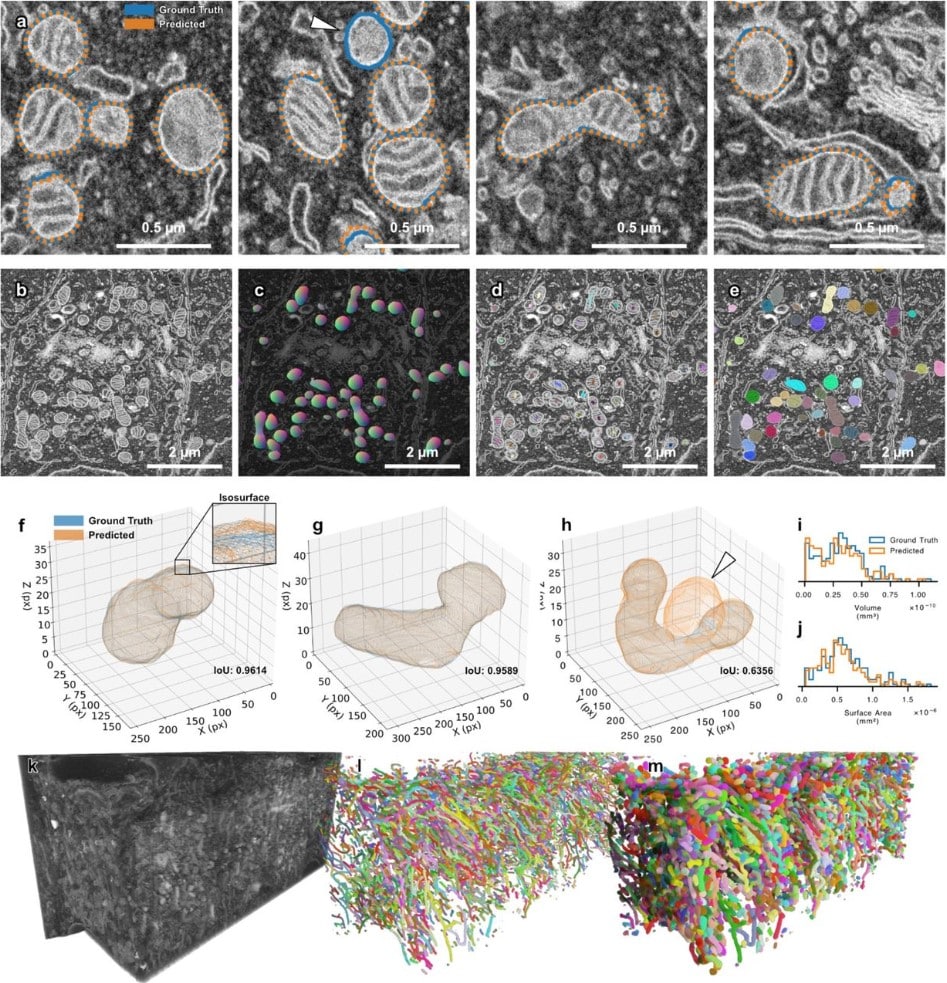
SKOOTS: Skeleton oriented object segmentation for mitochondria
Christopher J Buswinka, Hidetomi Nitta, Richard T. Osgood, Artur A. Indzhykulian
CLOOME: contrastive learning unlocks bioimaging databases for queries with chemical structures
Ana Sanchez-Fernandez, Elisabeth Rumetshofer, Sepp Hochreiter, Günter Klambauer
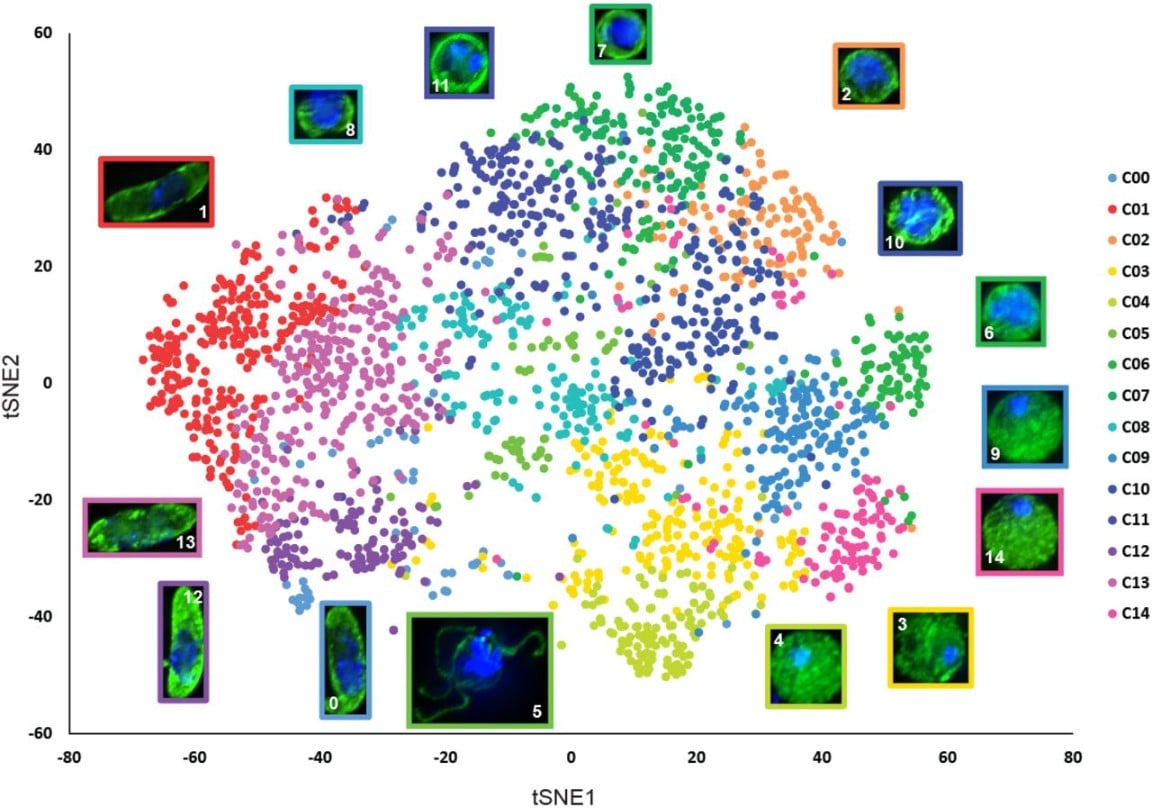
Machine Learning-based Phenotypic Imaging to Characterise the Targetable Biology of Plasmodium falciparum Male Gametocytes for the Development of Transmission-Blocking Antimalarials
Oleksiy Tsebriy, Andrii Khomiak, Celia Miguel-Blanco, Penny C. Sparkes, Maurizio Gioli, Marco Santelli, Edgar Whitley, Francisco-Javier Gamo, Michael J. Delves
WebAtlas pipeline for integrated single cell and spatial transcriptomic data
Tong Li, David Horsfall, Daniela Basurto-Lozada, Kenny Roberts, Martin Prete, John E G Lawrence, Peng He, Elisabeth Tuck, Josh Moore, Shila Ghazanfar, Sarah Teichmann, Muzlifah Haniffa, Omer Ali Bayraktar
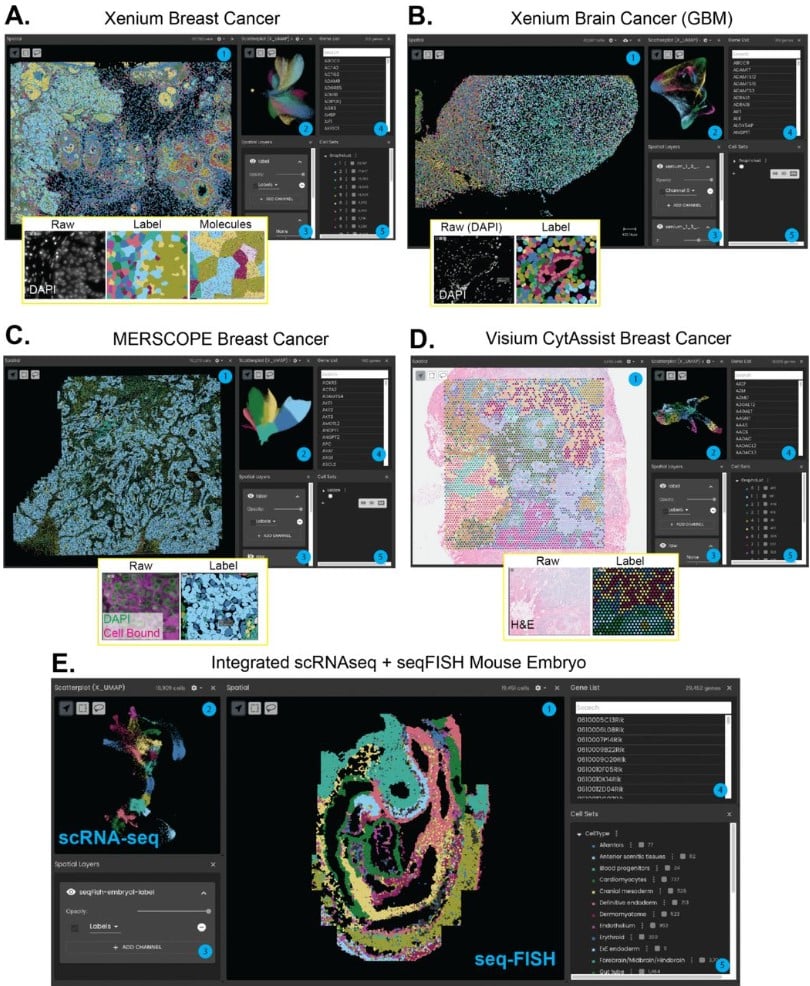
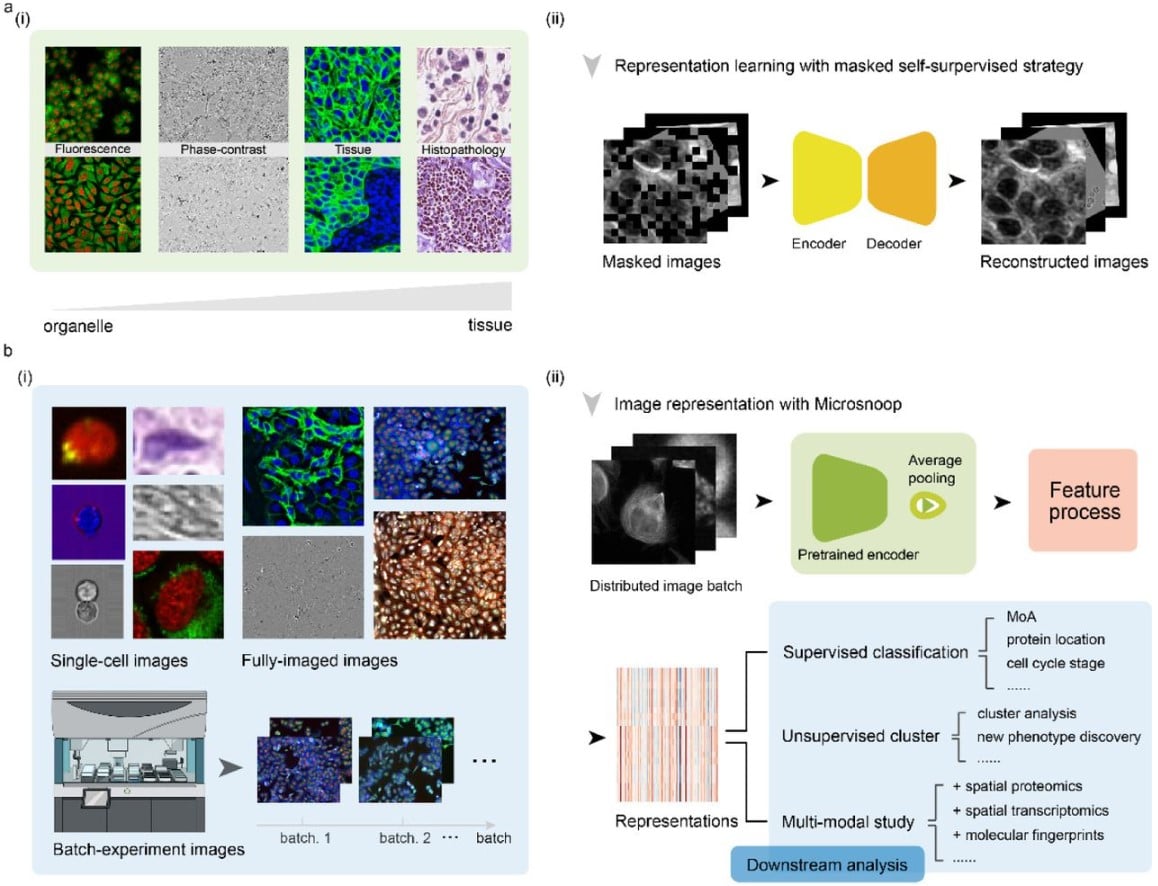
Microsnoop: A Generalized Tool for Unbiased Representation of Diverse Microscopy Images (v5)
Dejin Xun, Rui Wang, Xingcai Zhang, Yi Wang
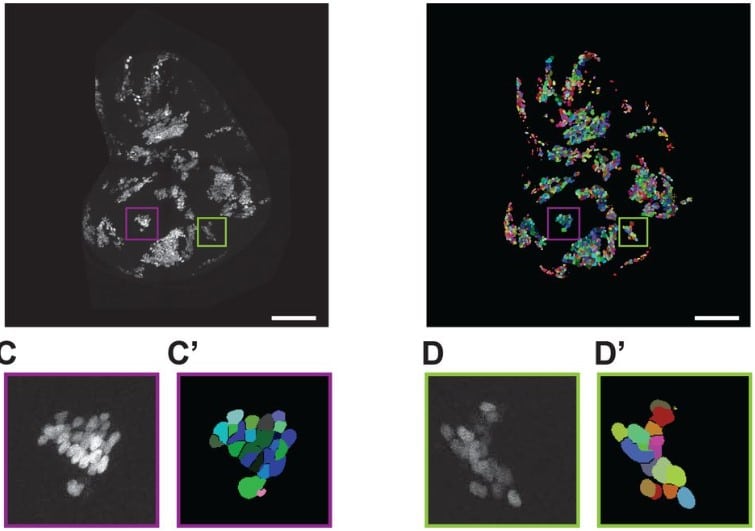
Automated counting of Drosophila imaginal disc cell nuclei
Pablo Sanchez Bosch, Jeffrey D. Axelrod
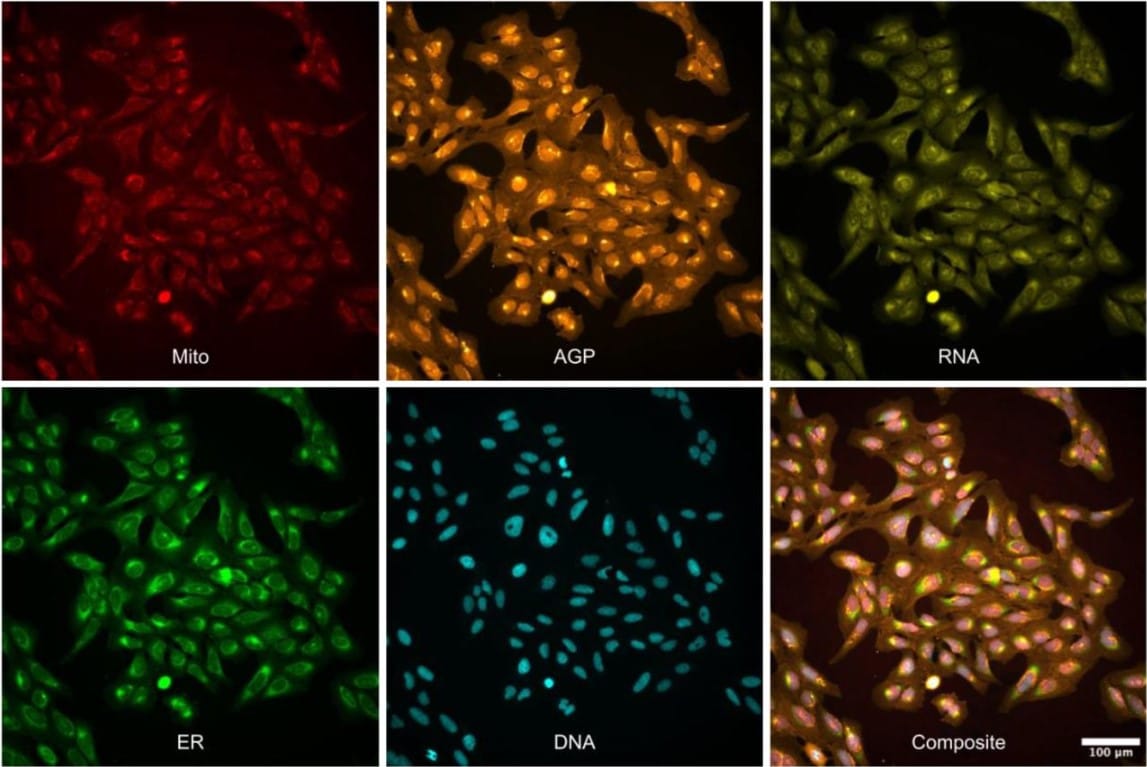
Three million images and morphological profiles of cells treated with matched chemical and genetic perturbations
Srinivas Niranj Chandrasekaran, Beth A. Cimini, Amy Goodale, Lisa Miller, Maria Kost-Alimova, Nasim Jamali, John G. Doench, Briana Fritchman, Adam Skepner, Michelle Melanson, John Arevalo, Marzieh Haghighi, Juan Caicedo, Daniel Kuhn, Desiree Hernandez, Jim Berstler, Hamdah Shafqat-Abbasi, David Root, Susanne E. Swalley, Sakshi Garg, Shantanu Singh, Anne E. Carpenter
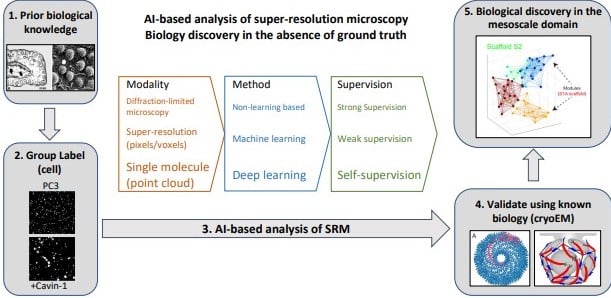
AI-based analysis of super-resolution microscopy: Biological discovery in the absence of ground truth
Ivan R. Nabi, Ben Cardoen, Ismail M. Khater, Guang Gao, Timothy H. Wong, Ghassan Hamarneh


 (No Ratings Yet)
(No Ratings Yet)