Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 29 December 2023
Here is a curated selection of preprints published recently. In this post, we focus specifically on new tools for bioimage analysis, released before 20 December.
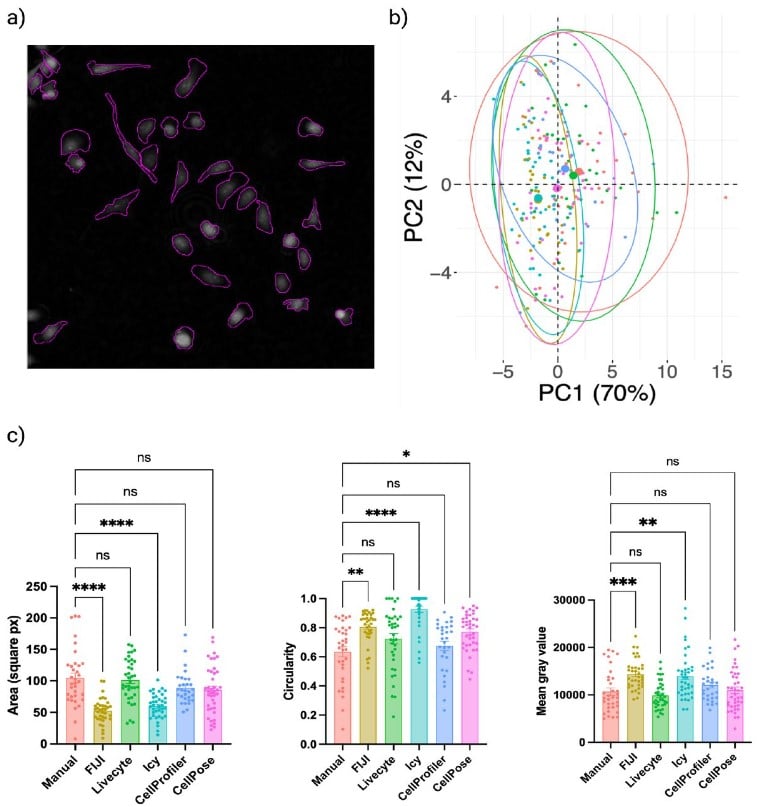
Exploring the impact of variability in cell segmentation and tracking approaches
Laura Wiggins, Peter J. O’Toole, William J. Brackenbury, Julie Wilson
EMformer: Transformer-Based Isotropic Reconstruction for Volume Electron Microscopy
Jia He, Yan Zhang, Wenhao Sun, Ge Yang, Fei Sun
NanoLocz: Image analysis platform for AFM, high-speed AFM and localization AFM
George R Heath, Emily Micklethwaite, Tabitha Storer
High-fidelity, high-resolution light-field reconstruction with physics-informed neural representation learning
Chengqiang Yi, Yifan Ma, Xinyue Yuan, Lanxin Zhu, Jiahao Sun, Shangbang Gao, Meng Zhang, Yuhui Zhang, Zhaoqiang Wang, Hsiai Tzung, Dongyu Li, Binbing Liu, Peng Fei
Fine-tuning TrailMap: The utility of transfer learning to improve the performance of deep learning in axon segmentation of light-sheet microscopy images
Marjolein Oostrom, Michael A. Muniak, Rogene M. Eichler West, Sarah Akers, Paritosh Pande, Moses Obiri, Wei Wang, Kasey Bowyer, Zhuhao Wu, Lisa M. Bramer, Tianyi Mao, Bobbie Jo Webb-Robertson
Towards Generalizability and Robustness in Biological Object Detection in Electron Microscopy Images
Katya Giannios, Abhishek Chaurasia, Cecilia Bueno, Jessica L. Riesterer, Lucas Pagano, Terence P. Lo, Guillaume Thibault, Joe W. Gray, Xubo Song, Bambi DeLaRosa
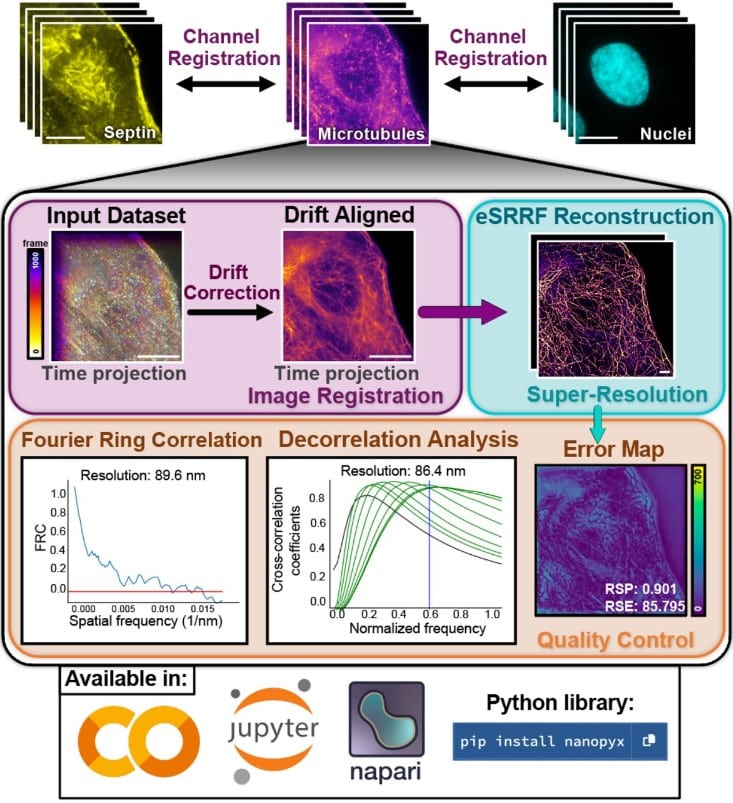
NanoPyx: super-fast bioimage analysis powered by adaptive machine learning
Bruno M. Saraiva, Inês M. Cunha, António D. Brito, Gautier Follain, Raquel Portela, Robert Haase, Pedro M. Pereira, Guillaume Jacquemet, Ricardo Henriques
VoltRon: A Spatial Omics Analysis Platform for Multi-Resolution and Multi-omics Integration using Image Registration
Artür Manukyan, Ella Bahry, Emanuel Wyler, Erik Becher, Anna Pascual-Reguant, Izabela Plumbom, Hasan Onur Dikmen, Sefer Elezkurtaj, Thomas Conrad, Janine Altmüller, Anja E. Hauser, Andreas Hocke, Helena Radbruch, Deborah Schmidt, Markus Landthaler, Altuna Akalin
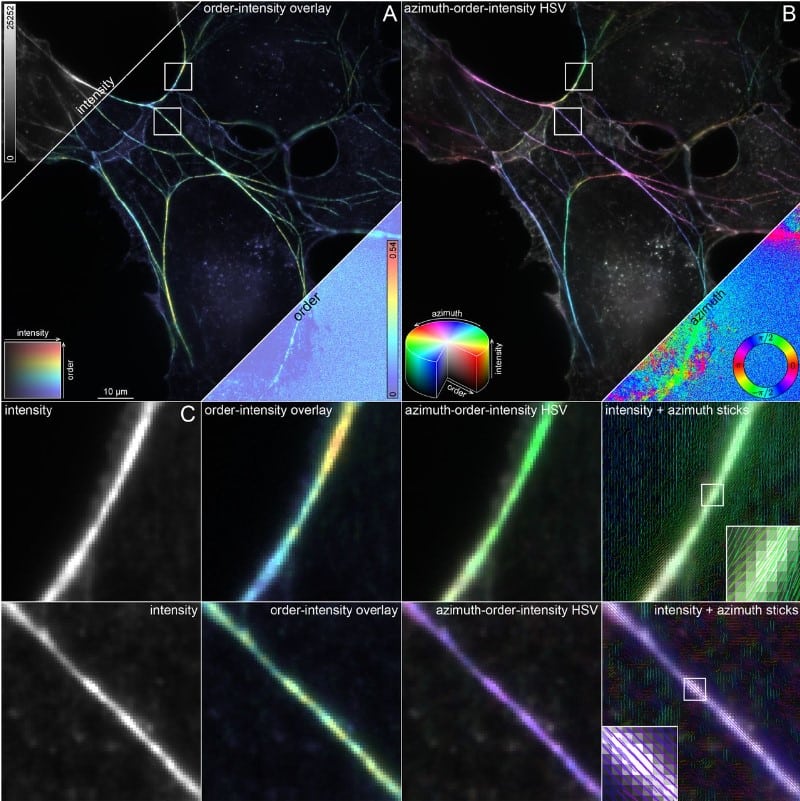
OOPS: Object-Oriented Polarization Software for analysis of fluorescence polarization microscopy images
William F. Dean, Tomasz J. Nawara, Rose M. Albert, Alexa L. Mattheyses
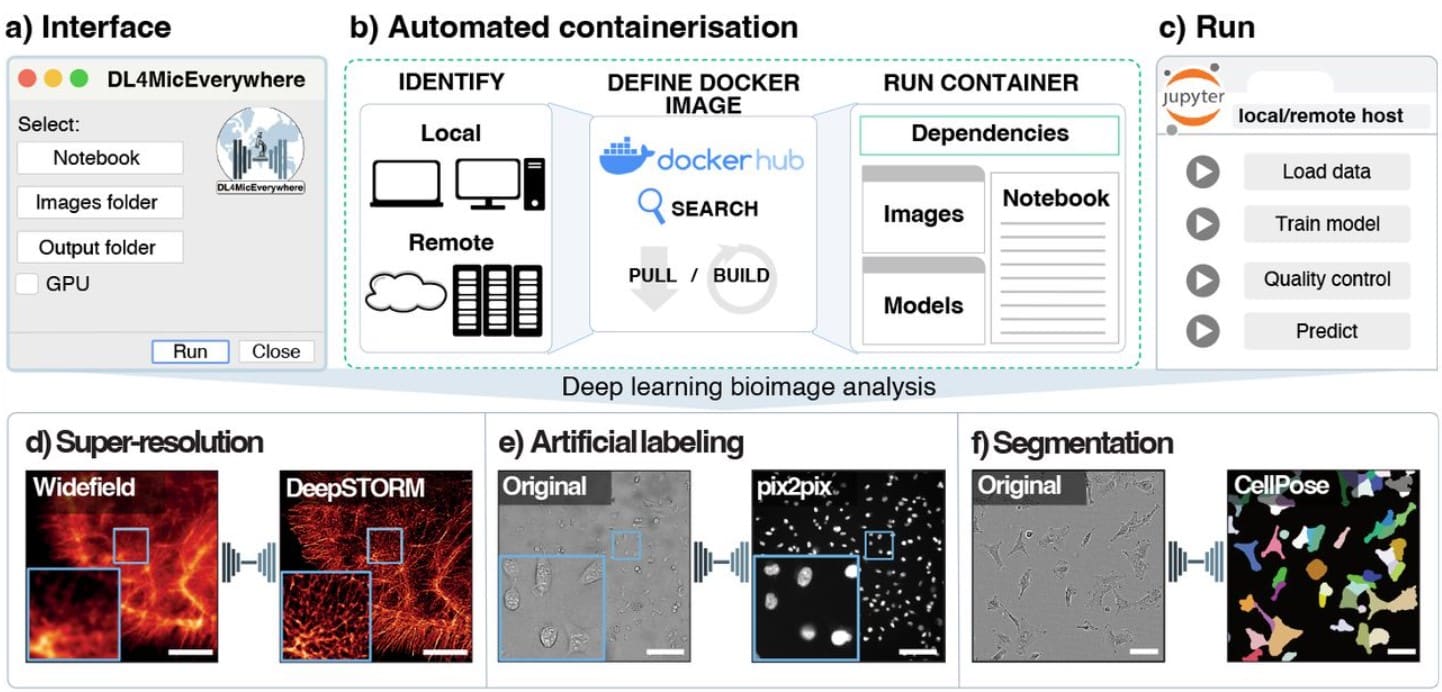
DL4MicEverywhere: Deep learning for microscopy made flexible, shareable, and reproducible
Iván Hidalgo-Cenalmor, Joanna W Pylvänäinen, Mariana G Ferreira, Craig T Russell, Ignacio Arganda-Carreras, AI4Life Consortium, Guillaume Jacquemet, Ricardo Henriques, Estibaliz Gómez-de-Mariscal
3D Nuclei Segmentation by Combining GAN Based Image Synthesis and Existing 3D Manual Annotations
Xareni Galindo, Thierno Barry, Pauline Guyot, Charlotte Rivière, Rémi Galland, Florian Levet
An arginine-rich nuclear localization signal (ArgiNLS) strategy for streamlined image segmentation of single-cells
Eric R. Szelenyi, Jovana S. Navarrete, Alexandria D. Murry, Yizhe Zhang, Kasey S. Girven, Lauren Kuo, Marcella M. Cline, Mollie X. Bernstein, Mariia Burdyniuk, Bryce Bowler, Nastacia L. Goodwin, Barbara Juarez, Larry S. Zweifel, Sam A. Golden
A benchmarked comparison of software packages for time-lapse image processing of monolayer bacterial population dynamics
Atiyeh Ahmadi, Matthew Courtney, Carolyn Ren, Brian Ingalls
Capturing cell heterogeneity in representations of cell populations for image-based profiling using contrastive learning
Robert van Dijk, John Arevalo, Mehrtash Babadi, Anne E. Carpenter, Shantanu Singh
blik: an extensible napari plugin for cryo-ET data visualisation, annotation and analysis
Lorenzo Gaifas, Joanna Timmins, Irina Gutsche
pHusion: A robust and versatile toolset for automated detection and analysis of exocytosis
Ellen C. O’Shaughnessy, Mable Lam, Samantha E. Ryken, Theresa Wiesner, Kimberly Lukasik, Bradley J Zuchero, Christophe Leterrier, David Adalsteinsson, Stephanie L. Gupton
Refinement of Cryo-EM 3D Maps with Self-Supervised Denoising Model: crefDenoiser
Ishaant Agarwal, Joanna Kaczmar-Michalska, Simon F. Nørrelykke, Andrzej J. Rzepiela
Unbiased Complete Estimation of Chloroplast Number in Plant Cells Using Deep Learning Methods
Qun Su, Le Liu, Zhengsheng Hu, Tao Wang, Huaying Wang, Qiuqi Guo, Xinyi Liao, Zhao Dong, Shaokai Yang, Ningjing Liu, Qiong Zhao
Deep learning assisted single particle tracking for automated correlation between diffusion and function
Jacob Kæstel-Hansen, Marilina de Sautu, Anand Saminathan, Gustavo Scanavachi, Ricardo F. Bango Da Cunha Correia, Annette Juma Nielsen, Sara Vogt Bleshøy, Wouter Boomsma, Tom Kirchhausen, Nikos S. Hatzakis
FluoroTensor: identification and tracking of colocalised molecules and their stoichiometries in multi-colour single molecule imaging via deep learning
Max F.K. Wills, Carlos Bueno Alejo, Nikolas Hundt, Andrew J. Hudson, Ian C. Eperon
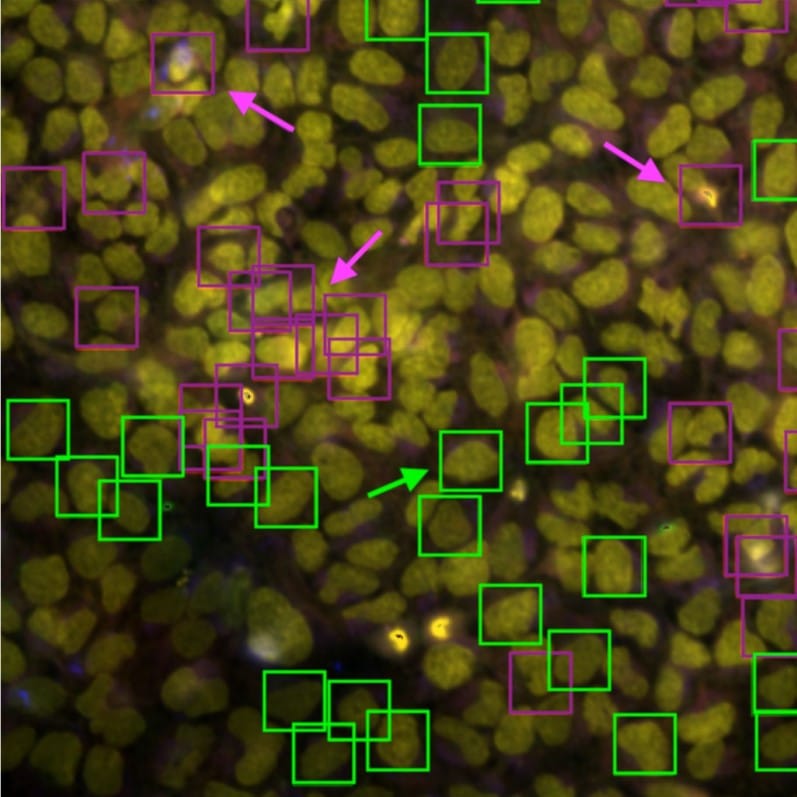
A Foundation Model for Cell Segmentation
Uriah Israel, Markus Marks, Rohit Dilip, Qilin Li, Morgan Schwartz, Elora Pradhan, Edward Pao, Shenyi Li, Alexander Pearson-Goulart, Pietro Perona, Georgia Gkioxari, Ross Barnowski, Yisong Yue, David Van Valen
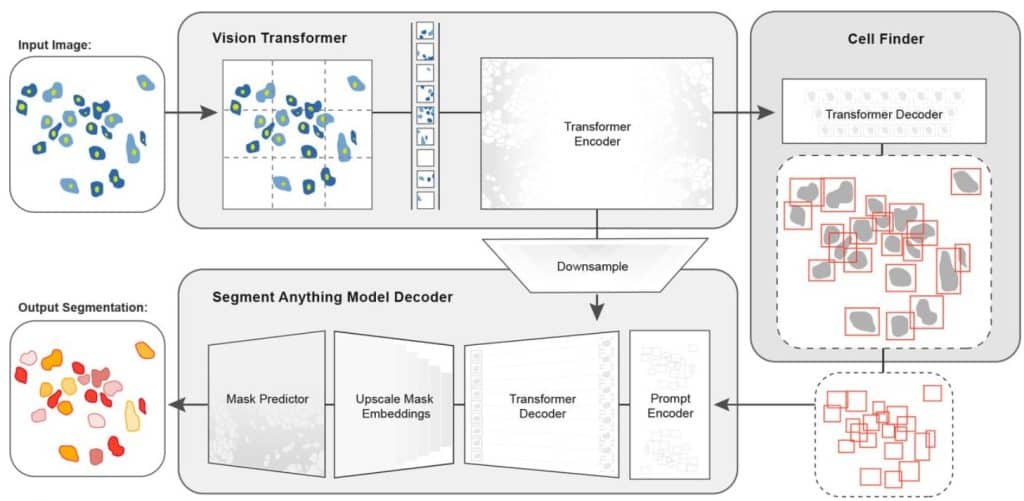
Figure extracted from Israel, et al. The image is made available under a CC-BY-4.0 International license.
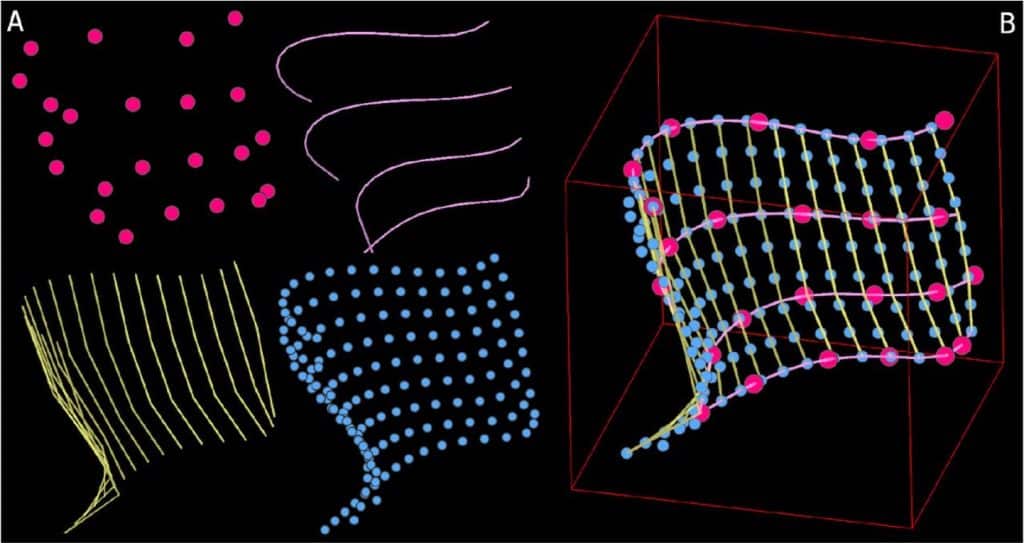
Automated cell annotation in multi-cell images using an improved CRF_ID algorithm
Hyun Jee Lee, Jingting Liang, Shivesh Chaudhary, Sihoon Moon, Zikai Yu, Taihong Wu, He Liu, Myung-Kyu Choi, Yun Zhang, Hang Lu
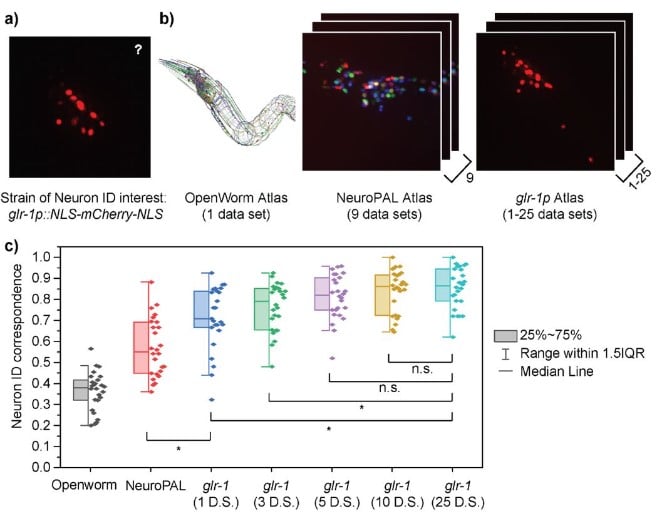
Figure extracted from Lee, et al. The image is made available under a CC-BY-4.0 International license.
Cell Spotter (CSPOT): A machine-learning approach to automated cell spotting and quantification of highly multiplexed tissue images
Ajit J. Nirmal, Clarence Yapp, Sandro Santagata, Peter K. Sorger
Bio-Image Informatics Index BIII: A unique database of image analysis tools and workflows for and by the bioimaging community
Chong Zhang, Alban Gaignard, Matus Kalas, Florian Levet, Felipe Delestro, Joakim Lindblad, Natasa Sladoje, Laure Plantard, Alain Latour, Robert Haase, Gabriel Martins, Paula Sampaio, Leandro Scholz, NEUBIAS taggers, Sébastien Tosi, Kota Miura, Julien Colombelli, Perrine Paul-Gilloteaux
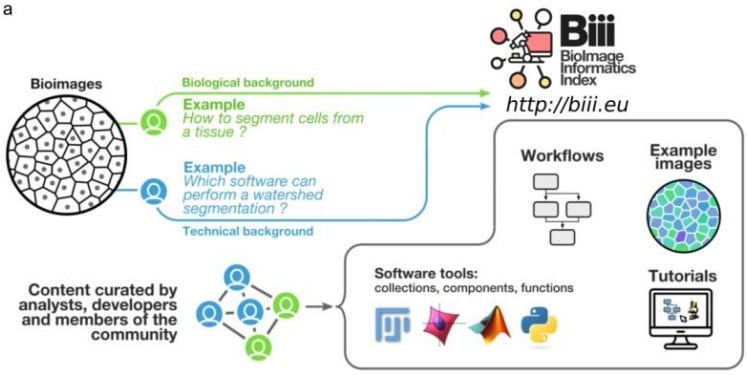
Nondestructive, quantitative viability analysis of 3D tissue cultures using machine learning image segmentation
Kylie J. Trettner, Jeremy Hsieh, Weikun Xiao, Jerry S.H. Lee, Andrea M. Armani
Fokker-Planck analysis of superresolution microscopy images
Mario Annunziato, Alfio Borzì
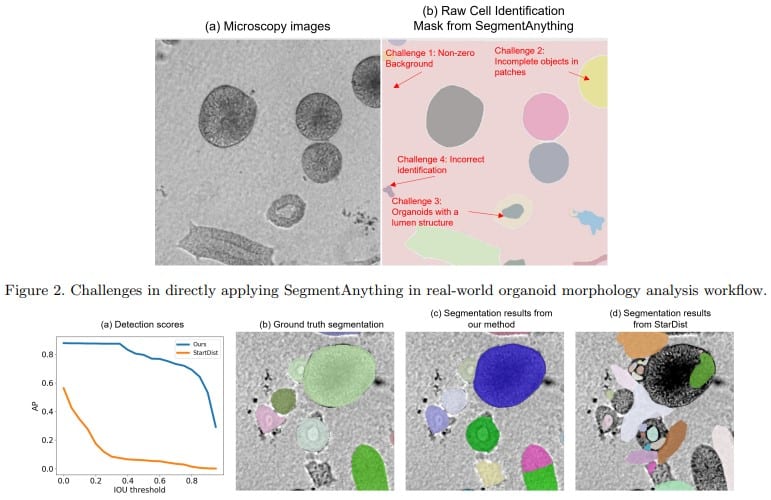
SegmentAnything helps microscopy images based automatic and quantitative organoid detection and analysis
Xiaodan Xing, Chunling Tang, Yunzhe Guo, Nicholas Kurniawan, Guang Yang
Gravitational cell detection and tracking in fluorescence microscopy data
Nikomidisz Eftimiu, Michal Kozubek
High-speed image reconstruction for nonlinear structured illumination microscopy
Jingxiang Zhang, Tianyu Zhao, Xiangda Fu, Manming Shu, Jiajing Yan, Jinxiao Chen, Yansheng Liang, Shaowei Wang, Ming Lei
Defining the boundaries: challenges and advances in identifying cells in microscopy images
Nodar Gogoberidze, Beth A. Cimini
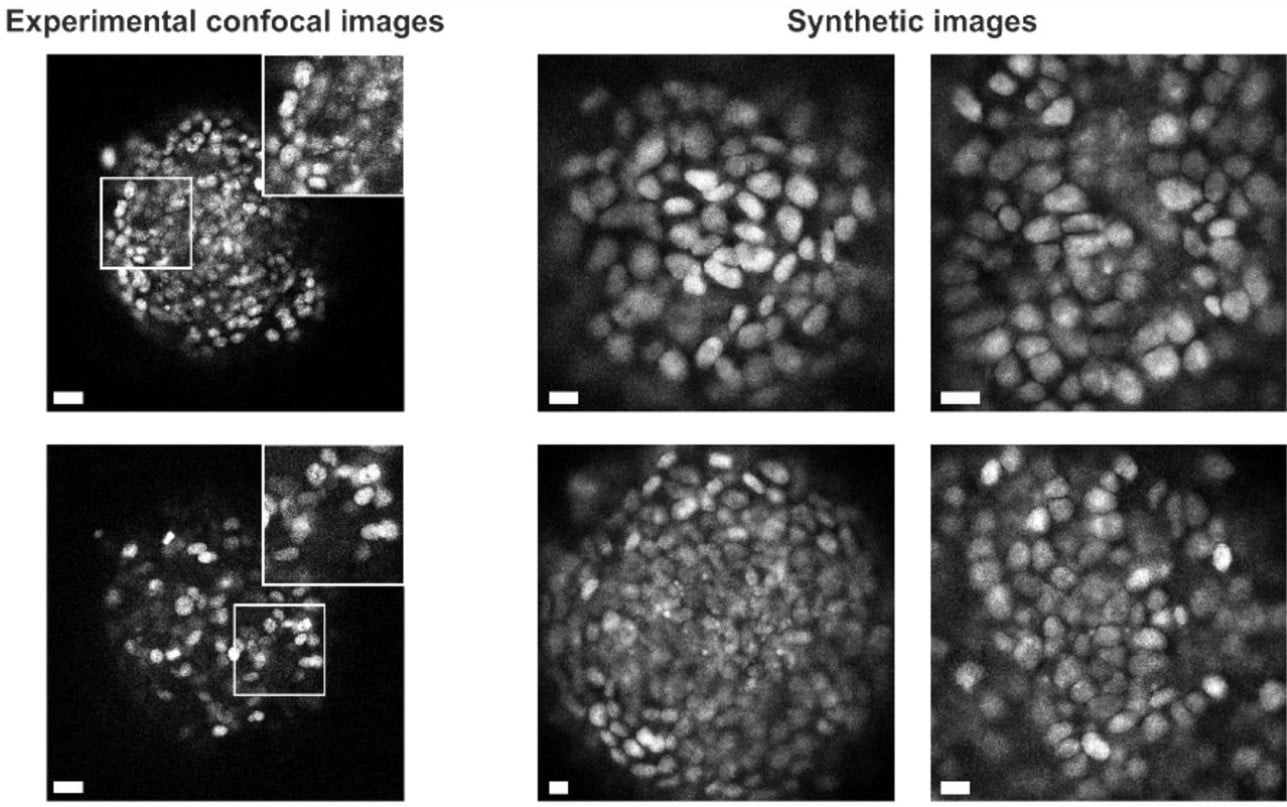
Microscopy Image Segmentation via Point and Shape Regularized Data Synthesis
Shijie Li, Mengwei Ren, Thomas Ach, Guido Gerig


 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)