If you can detect it, you can track it – recent TrackMate developments
Posted by Joanna Pylvänäinen, on 17 February 2022
TrackMate is your buddy for your everyday tracking
TrackMate (Tinevez et al., 2017) is an open-source Fiji plugin initially made for tracking single particles imaged with fluorescence microscopes. It has now been updated to include a collection of new detectors and direct implementation of deep-learning models (Ershov, Phan, Pylvänäinen, Rigaud et al., 2021). In addition to a variety of detectors and tracking algorithms, TrackMate includes advanced visualization and analysis tools, as well as powerful tools for track filtering and curation, giving us maximal control over the obtained results. TrackMate was created by Jean-Yves Tinevez, who hid the TrackMate code behind a nice, interactive and intuitive user interface. Before the update, TrackMate was already one of the most used and cited tracking tools for bioimage analysis. We can only dream about what the new and improved version of TrackMate will bring. Here is my story with TrackMate.
The story begins
My path through academia has been full of strange turns, although my interest in cell biology and user-friendly image analysis tools has always remained. I’ve tested TrackMate a few times in the past, but usually, I would stop using it mainly due to its limited ability to detect complex objects. Then I heard about my dear friend Elnaz Fazeli working with an image analysis workflow, where StarDist detection and cell tracking with TrackMate were combined in a user-friendly pipeline (Fazeli et al., 2020). I was very impressed. So no wonder, that when my Ph.D. supervisor Guillaume Jacquemet asked if I was interested in contributing to the new version of the TrackMate, I was like “Yeah – let’s do it!”. Although honestly, still at that point I did not yet fully understand the impact that this new TrackMate would have on the scientific community. Let’s take a look.
The new version of the TrackMate has improved object detection
The previous version of the TrackMate had excellent detectors for the detection of light blob-like objects on dark background using the Laplacian of Gaussian (LoG) or Difference of Gaussians (DoG) detectors. While these detectors perform well for fluorescent images, they still struggle when trying to detect textured, irregular objects, or objects imaged in transmitted-light microscopy (bright-field, phase contrast, DIC). As image segmentation, machine-learning (ML) and deep learning (DL) methods have taken a massive leap forward in the last few years, it was time to incorporate some of these features into TrackMate. To do this, Jean-Yves integrated a variety of new detectors (figure 1), such as detectors for label images, binary mask images and segmentation probability maps to enable detection of previously segmented objects. It is also possible to detect objects directly in the TrackMate user interface using the threshold detector and MorphoLibJ morphological segmentation algorithm. As a cherry on top of the cake, Jean-Yves incorporated DL and ML segmentation algorithms, such as StarDist, cellpose, ilastik and Weka to TrackMate, making it possible to apply custom-made segmentation algorithms to a specific dataset directly. The StarDist segmentation is one of my favorite segmentation algorithms, and needless to say, incorporation of that into TrackMate made me very happy! These segmentation improvements elevated TrackMate as a tracking tool to an entirely new level.
Here are some examples of the new detectors in action:
The new TrackMate has improved analysis capabilities
Another fantastic development of the new TrackMate is its improved analysis tools. The previous version of TrackMate was able to measure the objects’ position and tracks. Now with the new TrackMate, in addition to measuring the position of the objects, the integrated segmentation algorithms can provide additional information about the objects’ morphological features and even signal intensity in every frame. This allows us to correlate cells’ motility with changes in their shape and the fluorescent intensity over time. To showcase this feature, I generated a dataset, where I used a pre-trained StarDist model for nuclei segmentation in one channel and measured signal oscillation of an ERK-activity reporter in the segmented area over time in the other channel. We visualized the changes in ERK activity as heat maps using PlotTwist (https://huygens.science.uva.nl/) (Figure 2). A neat example application of morphological measurements during tracking is to correlate lineage information with the morphological measurements of objects, like was done in a study of Neisseria meningitidis growth (Figure 3).
The new TrackMate exports files for further analysis
I believe that there is no one correct way to analyze tracking data and not all the analysis can or should be done in TrackMate. What makes TrackMate extremely flexible is its versatile file exports that enable further analysis of the tracks using other platforms. For example, a useful way to further analyze cell tracks is to export them from TrackMate as a MotilityLab compatible spreadsheet and directly paste them to the MotilityLab online tool. MotilityLab (www.motilitylab.net, also called celltrackR) (Wortel et al., 2021) is an open-source R package containing several cell track analysis methods such as extracting and visualizing migration statistics, clustering tracks and simulating cell migration. MotilityLab can provide additional analysis methods, such as track straightness, that TrackMate cannot yet perform (Figure 4).
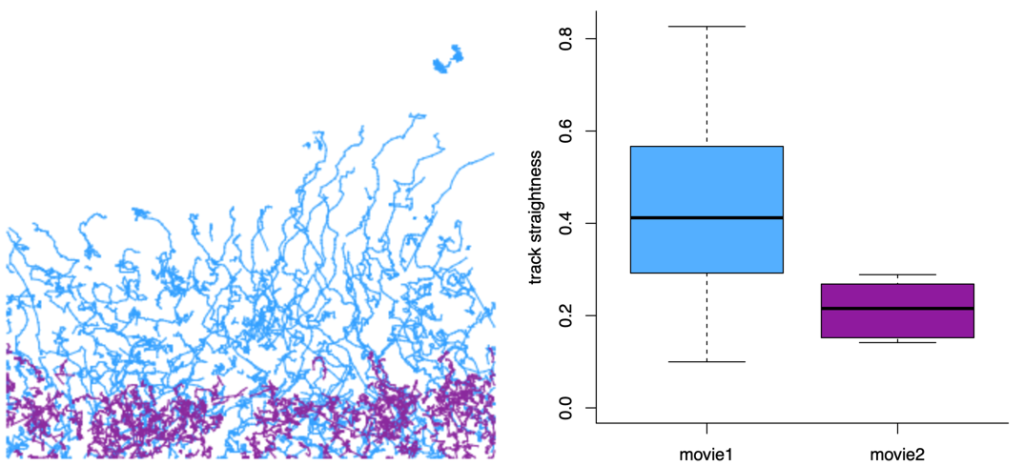
TrackMate can create 3D reconstructions
One of my favorite features of the New TrackMate is the possibility to create 3D label images. In this method, a 3D image stack is first converted into a time sequence (z -> t) before tracking. The objects can then be first detected using a suitable detector and then tracked. The tracking results are then exported as labels on tracks. After label image exportation, the time is swapped back to stack (t -> z) to transform the labels in time into 3D labels (video 3). This is a handy alternative for some of those currently existing time-consuming and computationally heavy 3D segmentation solutions.
Availability and tutorials
The new TrackMate is super easy to install, it is available through multiple Fiji update sites and is extensively documented. The documentation regarding all new features can be found here: https://imagej.net/plugins/TrackMate/TrackMate-v7-detectors. Additionally, you can look for the best possible detector for your use case using the cheat sheet poster below (Figure 5).
Final words
The new version of TrackMate has already revolutionized how we can detect and track our imaging data, making it possible to track almost anything. Overall, these new features make the new TrackMate your very best buddy for your everyday tracking. The story of TrackMate does not end here, it will be continuously developed and improved. To support this development, let’s all continue making great science using the new TrackMate. I know I will!
Acknowledgments
TrackMate was created by Jean-Yves Tinevez from the Institut Pasteur, Paris. Not only did he make the TrackMate from good to great, but he and his team have been a pleasure to work with. I want to also acknowledge Guillaume Jacquemet, who brought the biologist to the equation and pushed TrackMate to be as user-friendly as possible for all of us to use. Thanks for having me on board!

posted by
Joanna Pylvänäinen
Image data coordinator at Turku BioImaging
PhD student at Guillaume Jacquemet Lab
Åbo Akademi University, Turku, Finland
(picture by Solveig Eriksson)
Literature
Ershov, D., Phan, M.-S., Pylvänäinen, J.W., Rigaud, S.U., Le Blanc, L., Charles-Orszag, A., Conway, J.R.W., Laine, R.F., Roy, N.H., Bonazzi, D., Duménil, G., Jacquemet, G., Tinevez, J.-Y., 2021. Bringing TrackMate in the era of machine-learning and deep-learning. bioRxiv 2021.09.03.458852. https://doi.org/10.1101/2021.09.03.458852
Fantham, M., Kaminski, C.F., 2017. A new online tool for visualization of volumetric data. Nat. Photonics 11, 69–69. https://doi.org/10.1038/nphoton.2016.273
Fazeli, E., Roy, N.H., Follain, G., Laine, R.F., von Chamier, L., Hänninen, P.E., Eriksson, J.E., Tinevez, J.-Y., Jacquemet, G., 2020. Automated cell tracking using StarDist and TrackMate. F1000Research 9, 1279. https://doi.org/10.12688/f1000research.27019.1
Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G.M., Reynolds, G.D., Laplantine, E., Bednarek, S.Y., Shorte, S.L., Eliceiri, K.W., 2017. TrackMate: An open and extensible platform for single-particle tracking. Methods San Diego Calif 115, 80–90. https://doi.org/10.1016/j.ymeth.2016.09.016
von Chamier, L., Laine, R.F., Jukkala, J., Spahn, C., Krentzel, D., Nehme, E., Lerche, M., Hernández-Pérez, S., Mattila, P.K., Karinou, E., Holden, S., Solak, A.C., Krull, A., Buchholz, T.-O., Jones, M.L., Royer, L.A., Leterrier, C., Shechtman, Y., Jug, F., Heilemann, M., Jacquemet, G., Henriques, R., 2021. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. 12, 2276. https://doi.org/10.1038/s41467-021-22518-0
Wortel, I.M.N., Liu, A.Y., Dannenberg, K., Berry, J.C., Miller, M.J., Textor, J., 2021. CelltrackR: An R package for fast and flexible analysis of immune cell migration data. ImmunoInformatics 1. https://doi.org/10.1016/j.immuno.2021.100003


 (3 votes, average: 1.00 out of 1)
(3 votes, average: 1.00 out of 1)
