Expansion microscopy
Posted by Shoh Asano, on 29 July 2020
Written by Shoh Asano and Ruixuan Gao
Light microscopy and diffraction limit
For centuries, light microscopy has played a central role in biological studies. The first implementations of a light microscope dates back to as early as the late 16th and early 17th century, when an array of polished lenses was used to magnify (biological) objects to reveal intricate details no one had seen before. While the basic concept of microscopy has been applied and implemented in a wide range of scientific fields, the importance of light microscopy to current biological research cannot be overstated – some 400 years after its introduction.
One of the most important improvements in light microscopy came with the introduction of fluorescent tags. With the discovery and synthesis of fluorescent proteins (that can be produced within cells or organisms) and fluorescent dyes, specific structures or molecules in cells and tissues could be fluorescently labeled. When illuminated, these fluorescently-tagged molecules will absorb light of a certain wavelength, and irradiate (or fluoresce) back at a different wavelength. Named fluorescence microscopy, this technique has become a powerful tool for modern biological research.
Fluorescence microscopy produced countless biological and medical discoveries far beyond the scope of this article, but had a fundamental, and physical limit to its resolution. At the end of the 19th century, Ernst Abbe proposed that an optical system would not be able to tell apart two sources of light beyond a certain limit due to diffraction of light waves. As a result, conventional widefield fluorescence microscopy cannot resolve structures smaller than ~200 nanometers, the diffraction-limit at its shortest emission wavelength.
For any structural studies beyond this limit, other illumination sources were required, such as electrons or x-rays. However, compared to these techniques, fluorescence microscopy offers considerable advantages in sample preservation, usability, molecular specificity, and temporal resolution. For example, fluorescence microscopy can capture multi-color images of tagged cells and tissues that contain molecular-specific information (e.g. using fluorescent tags of different emission wavelengths). Moreover, it is relatively simple to operate a fluorescence microscope and prepare the fluorescently-tagged samples (compared to other higher-resolution imaging techniques).
To close the gap between these technologies, many methods were developed in the last few decades to increase the resolution of fluorescence microscopy. This field is referred to as super-resolution microscopy, as it appears to defy Abbe’s law. Significant improvements in image resolution were enabled through switching (or modulating) the fluorescent emission – either by deterministic (or targeted) on/off switching of fluorescent molecules (e.g., stimulated emission depletion microscopy, STED) or stochastic on/off switching and a downstream image reconstruction process (e.g. photoactivated localization microscopy, PALM; or stochastic optical reconstruction microscopy, STORM). These techniques now allow features of down to few tens of nanometers to be resolved, and has been recently awarded the Nobel Prize in Chemistry.
Installing such a super-resolution imaging system, however, can be costly and technically challenging. Recently, an interesting concept to enhance resolution of fluorescence microscopy was proposed by Ed Boyden’s lab [1]. Termed expansion microscopy, the enhancement of resolution was generated by physical expansion of the sample itself, instead of the microscope design and/or the computational algorithms. This approach allowed individuals with access to diffraction-limit fluorescence microscopes to image biological specimens at higher effective resolution with minimal time and technical investment.
Physical expansion of biological specimens
In expansion microscopy, samples such as cells or tissues are embedded in a hydrogel and physically expanded (Fig. 1). The effective resolution is increased by making the sample larger, and thus the enhancement does not rely on any optical or algorithmic improvements. The initially demonstrated expansion factor was ~4.5-fold, which stipulates that a cell of 10 x 10 µm dimensions would turn into a 45 x 45 µm sample [1]. Using a conventional, diffraction-limited fluorescence microscopy setup that resolves biological details down to ~300nm, biological structures of ~70 nm (= 300 nm/4.5) in the original ‘unexpanded’ sample can now be resolved. This corresponds to a significant (4.5-fold) gain in the effective resolution.
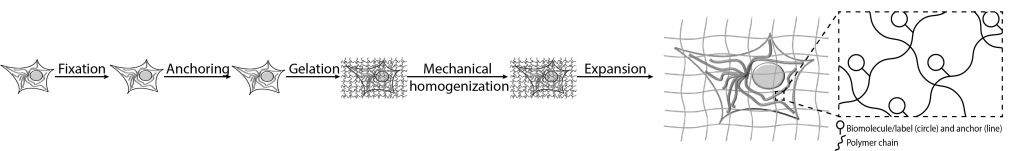
From a sample preparation perspective, expansion microscopy works with fixed biological specimens, ranging from cells, to tissue slices, and to an entire organism [2,3,4]. These samples can be either genetically tagged or immunostained – to mark the biomolecules of interest. A gel network is then formed in and around the samples, which serves as a matrix to anchor the target biomolecules, and to physically separate them in the final expansion step. By using specific chemical linkers, target biomolecules can be linked to this hydrogel network. After the gel is formed throughout the sample, the tissue is chemically or enzymatically digested to enable uniform expansion at molecular level. The digestion step can be carried out by several methods, such as through enzymes (e.g. proteinase K), or by denaturation (e.g. under high heat and/or pressure). After this step, the sample-gel composite can be immersed in water, where it physically swells. A similar chemistry and expansion process can be observed in super-absorbent materials, such as those used in diapers, when they come in contact with water.
Expansion microscopy offers additional benefits on top of the improved resolution. First, the expanded samples are optically clear. The transparency allows imaging of thicker specimens that conventional light microscopy could not image. Second, by physically expanding and thus ‘pulling apart’ the specimens, molecularly dense areas become more accessible to staining and/or amplification reagents. This decrowding effect is especially useful for localization applications described later.
Apart from these benefits, some caveats should also be noted for expansion microscopy. It only works with fixed samples. Thus, it precludes live sample imaging at effective super-resolution. Also, expanded samples are larger. In fact, a ~4.5-fold expansion would correspond to a close to ~100-fold volume increase (which increases the imaging volume by the same volumetric factor). This also means that the expanded sample is usually dimmer than the original sample, as the signal is spread across a larger volume. For imaging these large areas or volumes, it is therefore recommended to use high-speed, laser-based fluorescence microscopes, such as a spinning-disk confocal microscope or a light-sheet fluorescence microscope. Combining expansion microscopy with these microscopes would cover large field-of-views with minimal imaging time, and thus can be favorable to other super-resolution techniques in terms of imaging speed and photobleaching.
Development and application of expansion microscopy
Expansion microscopy is an emerging field. Since its introduction in 2015, there have been continuous developments and improvements. In the most commonly used expansion protocol, proteins of interest are directly linked to the hydrogel network [5,6]. Termed protein-retention expansion microscopy (proExM), the new protocol can be performed with off-the-shelf chemicals and simple lab equipment, which is far easier to implement than the initial protocol, which required in-house preparation of oligo-conjugated antibodies. Furthermore, new protocols with highly effective fixation and staining strategies, such as expansion passive CLARITY technique (ePACT) [7] and magnified analysis of the proteome (MAP) [8], enabled tissue expansion at whole-organ or whole-body scale.
In the emerging field of spatial analysis of RNA transcripts, expansion microscopy can be utilized to carry out fluorescence in situ hybridization imaging (ExFISH) [9] and in situ sequencing (ExSeq) [10] of RNAs at nanoscopic resolution. The previously described “decrowding” effect of otherwise closely positioned RNAs provides more space for sequencing and amplification chemistry, and thus ensures a higher accuracy in fluorescent readouts and counts.
More recently, higher expansion factors of ~20-fold and beyond were achieved by iteratively expanding the specimens (iExM) [11]. In addition, an expansion factor of ~10x was demonstrated for single-round expansion [12]. These variants of expansion microscopy are particularly suitable for dense biological structures, such as lipid-stained cells and tissues [13]. Another version of expansion microscopy allows nanoscale imaging of clinical and pathological samples (e.g. human specimens) [14]. On the chemistry side, a new hydrogel matrix has been introduced to enable expansion (e.g. of viral particles) at sub-10 nm accuracy [15].
Finally, numerous improvements have been made to allow expansion microscopy to work across a diverse set of samples. To name a few, the technique has been applied to brain tissues, cell cultures, histological slices, mounted pathological samples, fly brains, and an entire C. elegans [1,3,4,10] (Fig. 2). A curated list of papers and protocols is available from the Boyden group.
Expansion microscopy will continue to evolve in myriad directions, such as with regard to its hydrogel chemistry and composition, linking chemistry, as well as the digestion condition. However, developments in other fields, including brighter and photostable fluorescent dyes and proteins, microscopes with fast imaging speed and low photobleaching, and robust image processing pipelines, will have direct impacts on the effectiveness and usability of expansion microscopy.
Looking ahead, expansion microscopy will be pivotal to visualize and trace large cellular morphologies and molecular constituents across a whole organism or multiple organisms with molecular information and nanoscopic resolution. Initial demonstration on an entire fly brain looks promising, and with aforementioned improvements, may be able to scale up to larger organisms [3]. Current and future imaging initiatives that require visualization and identification of molecular markers will likely benefit from expansion microscopy, such as brain connectomics (tracing each brain cell and its connections) and spatial proteomics and/or transcriptomics research. Finally, with ever-increasing CPU and GPU processing power, more advanced image processing algorithms and machine learning-based analyses, expansion microscopy datasets of substantially larger sizes can be acquired and processed routinely for biological experiments.
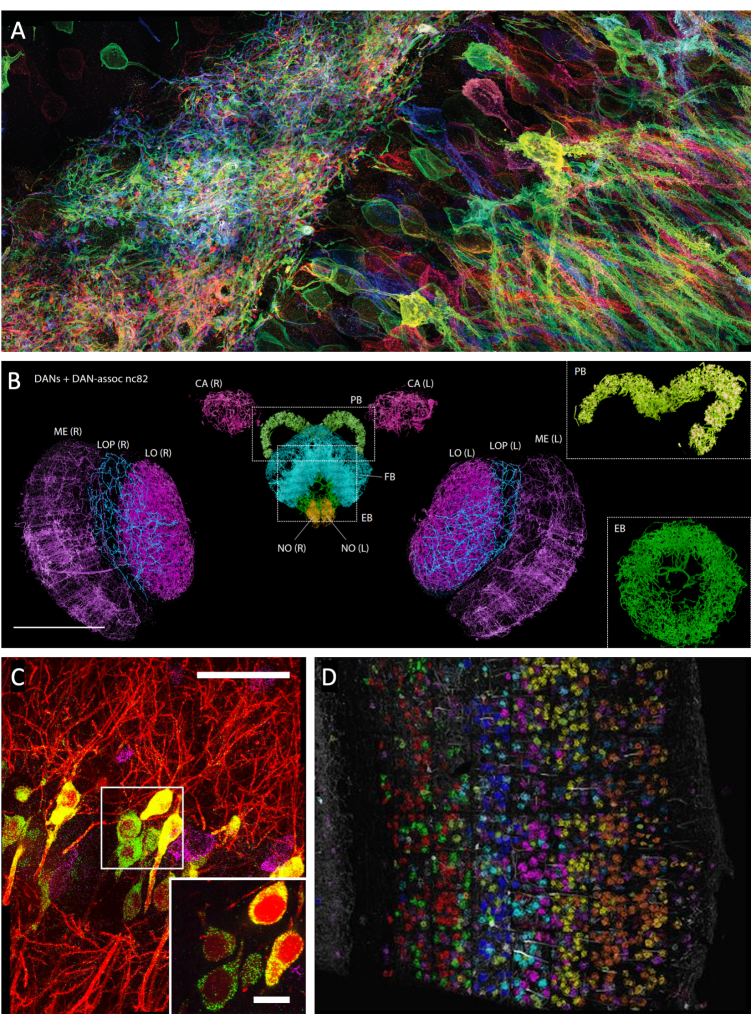
Figure 2. Nanoscale imaging of proteins and RNAs with expansion microscopy. (A) Membrane-labeled Brainbow 3.0 mouse hippocampus. Imaged with protein-retention expansion microscopy (proExM). (B) Dopaminergic neurons (DANs) and DAN-associated pre-synaptic sites (nc82, white puncta) in distinctive regions (colored) of an entire fly brain. Imaged with expansion lattice light-sheet microscopy (ExLLSM). Insets, protocerebral bridge (PB, top, angled view) and ellipsoid body (EB, bottom). Scale bar, 100 μm. (C) Thy1-YFP mouse hippocampus with single RNA puncta (red, YFP protein; cyan, YFP mRNA; magenta, Gad1 mRNA). Imaged with expansion FISH (ExFISH). Inset, one plane of the boxed region. Scale bar, 50 μm (inset: 10 μm). (D) Thy1-YFP mouse primary visual cortex with cell-segmented sequencing reads, colored by cluster assignment and overlaid on the YFP morphological marker (white). Sequenced and imaged with expansion sequencing (ExSeq). The figure is adapted and modified from (Alon et al., 2020; Chen et al., 2016; Gao et al., 2019b; Tillberg et al., 2016). [10, 9, 3, 6]

Shoh Asano
Principal Scientist
Internal Medicine Research Unit, Pfizer
USA

Ruixuan Gao
Postdoctoral Associate
Ed Boyden Lab, MIT
Cambridge, MA, USA


 (3 votes, average: 1.00 out of 1)
(3 votes, average: 1.00 out of 1)