Technology highlights – Structured Illumination Microscopy (SIM)
Posted by Johanna Bischof, on 30 September 2020
Interview with Ivan Novotný, Ph.D., from the Light Microscopy Core facility at the Institute of Molecular Genetics, in Prague, Czech Republic.
Please tell us a bit about yourself and the facility where you work.
I am employed as an imaging specialist at the Light Microscopy Core facility at the Institute of Molecular Genetics of the Czech Academy of Science in Prague, CZ. This facility is part of the Prague Node of Euro-BioImaging. I am really interested in different super-resolution (SR) methods, particularly Structured Illumination Microscopy (SIM).

We are today talking about Structured Illumination Microscopy. Can you please tell us about how SIM works?
As any other super-resolution method SIM aims to obtain multi-color fluorescence images at spatial resolution better than the physical limit given by light diffraction. I would start with the great benefits of SIM – the method does not require any special sample preparation and is relatively fast. The sample preparation procedure should be precise, as you want to see more than with a standard microscope, however, the fluorophores, sample fixation, mounting etc. you can keep from your standard protocol.
Moreover, the method is suitable for live cell imaging using standard expression vectors like EGFP to tag the protein of interest. Because we are talking about SR methods, we think about visualizing intracellular structures or generally structures that are much smaller than a typical cell. SIM is a camera-based microscope, which uses a moving structured excitation light pattern to excite fluorophores in the sample instead of the homogeneous illumination typical for standard wide-field fluorescence systems. For one SR image it is necessary to acquire 15 structurally illuminated images, which takes less than a second for a well stained sample. The method then uses a mathematical reconstruction in Fourier space to calculate the final SR image from the set of raw structurally illuminated images. Of course, both the acquisition and the reconstruction is automated and implemented in a user-friendly fashion in the microscope software.
In practice, we can expect to reach resolution around 120 nm in the green channel (emission at 520 nm) and around 150 nm in the red channel (emission at 670 nm). The method is linear, thus shorter wavelengths give a better resolution than the longer ones.
One could argue that a confocal scanning microscope with a pinhole closed to lower than 0.6 AU followed by deconvolution would also give you almost twice the resolution than a standard wide-field fluorescence microscope. But compared to that technique, SIM has the great advantage of acquiring a high amount of photons in one snapshot of the camera, which results in high contrast and lots of gathered information. At the same time, the speed of the acquisition with a spatial light modulator-based advanced system like our OMX V4 with Blaze module is closer to a spinning disc system than to the scanning confocal.
That sounds like SIM is a very powerful method. What kind of questions can be addressed using SIM?
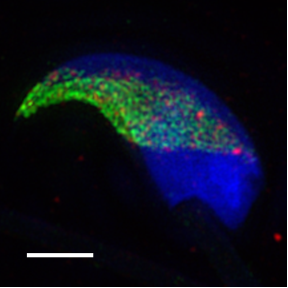
Author: Dr. Michaela Frolikova, Laboratory of Reproductive Biology, Institute of Biotechnology of the CAS, BIOCEV
The method serves to obtain SR images from a variety of standard fixed samples e.g. centromeres and nuclear pores where visualized with the method. We routinely use SIM for imaging anything from adherent cells, plant cells, mouse sperm cells (as you can see on the right here) and bacteria to thin tissue sections.
For live cell imaging, it is typically used for imaging microtubules, mitochondria and intra-nuclear non-membranous organelles like Cajal bodies.
A few examples of papers using SIM:
- The torpedo effect in Bacillus subtilis: RNase J1 resolves stalled transcription complexes [1]
- Chromatin organization at the nuclear periphery as revealed by image analysis of structured illumination microscopy data [2]
- SART3-Dependent Accumulation of Incomplete Spliceosomal snRNPs in Cajal Bodies [3]
Here’s a representative image of the bacterium Bacillus subtilis, that was collected using SIM at our facility:
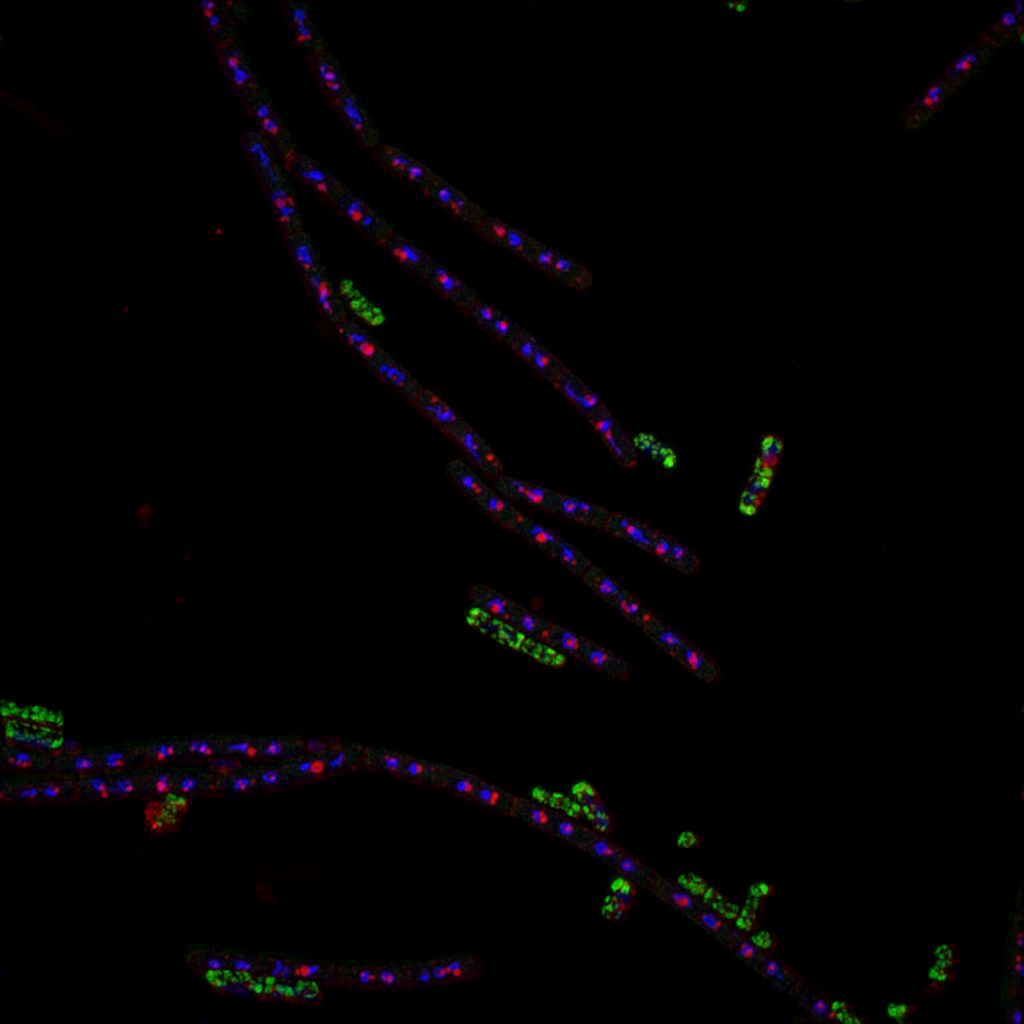
Author: Michaela Sikova, Laboratory of Microbial Genetics and Gene Expression, Institute of Microbiology of the CAS, CZ
Why is this technology best suited for looking at Bacillus?
In the particular case of Bacillus subtilis imaging, the user was limited to using standard staining protocols, but at the same time needed higher resolution in a large area for precise quantification of molecular distribution within small bacteria. EGFP expression, together with Nile red staining and DAPI counter-staining was used. In this case it was also important that the studied sample was nicely transparent – not thick and optically flat. The resolution of the resulting image using SIM is double that which is achievable using standard confocal imaging.
What are some challenges of this kind of microscopy? What do researchers have to pay attention to when performing these experiments?
It is important to mention also some limitations of the method. SIM is based on structured illumination of the sample and proper structured illumination is crucial for the reconstruction of the final SR image. The structured illumination can be affected by several things which we should consider before we choose this method. First, the spherical aberration introduces a smearing and blurring of the illumination pattern. Second, sample thickness can complicate achieving proper structured illumination and thus we do not recommend using SIM for samples thicker than 15 um. During acquisition, the user should check for bleaching within the acquired z-stack as bleaching can introduce serious artifacts to the final image.
When we stick to these recommendations, we can obtain very nice, clear and contrast-rich images with 2x higher resolution than a standard fluorescence microscope (according to the Abbe theorem).
What other services do you provide in your facility that would be useful in combination with this type of microscopy?
We usually provide our users with image analysis assistance when some quantitative or qualitative output is required. We are also advanced in sample preparation and many times we provide our users with advice and supervision during sample preparation steps. We also produce our own mounting media suitable for SR microscopy due to their optimized properties – anti-fade effect and a high refractive index. In some cases, when spatial resolution offered by SIM is not sufficient, we offer also STED and SMLM technologies, provided that the sample can be prepared according to the requirements of these methods.
To sum up, in our SR microscopy facility we help our users to establish the complete workflow – from proper sample preparation, followed by imaging, and finalized with image analysis.
Why should scientists choose your facility for using this technology?
We are quite familiar with the method (we provide the technology since 2015 as a service and I have practical experience with SIM since 2012) and we can supervise not only the imaging but also the sample preparation and help with image analysis. Our instrumentation for SIM is the DeltaVision OMX V4 with blaze module which provide us with a very stable and fast system suitable not only for fixed samples but also for live cell imaging.
Want to use SIM microscopy via the Euro-BioImaging service?
Euro-BioImaging is the European landmark research infrastructure for biological and biomedical imaging as recognised by the European Strategy Forum on Research Infrastructures (ESFRI). All scientists, regardless of their affiliation, area of expertise or field of activity can benefit from Euro-BioImaging’s pan-European open access services. By facilitating user access to high quality imaging facilities, resources, and services, with a constantly evolving technology offer, Euro-BioImaging will boost the productivity and impact of research across Europe.
Potential users of the Euro-BioImaging technologies are encouraged to submit project proposals via our website. To do so, you can LogIn to access our application platform, choose the technology you want to use and the facility you wish to visit, then submit your proposal. By using Euro-BioImaging, users will benefit from advice and guidance by technical experts working at the Nodes, training opportunities, and data management services.
For more information visit our website at www.eurobioimaging.eu or contact us at info@eurobioimaging.eu




 (No Ratings Yet)
(No Ratings Yet)