How I got into the Flamingo project, a new way of sharing custom-built light sheet microscopes
Posted by Michael Weber, on 12 October 2020
Early into my undergraduate studies, I started working as a student helper in Kurt Anderson’s (and later Jan Peychl’s) light microscopy facility (LMF) at the Max-Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG) in Dresden, Germany. Maintaining commercial microscope setups and helping researchers getting the best possible image data from their samples would eventually become a full-time job for several years. I even got to oversee my own little facility (now CFCI) with two confocal microscopes. All these commercial microscopes would provide great images to a certain extent, but I kept wondering how much one could push the boundaries of what’s possible if they would be more open and customizable for the user’s needs.
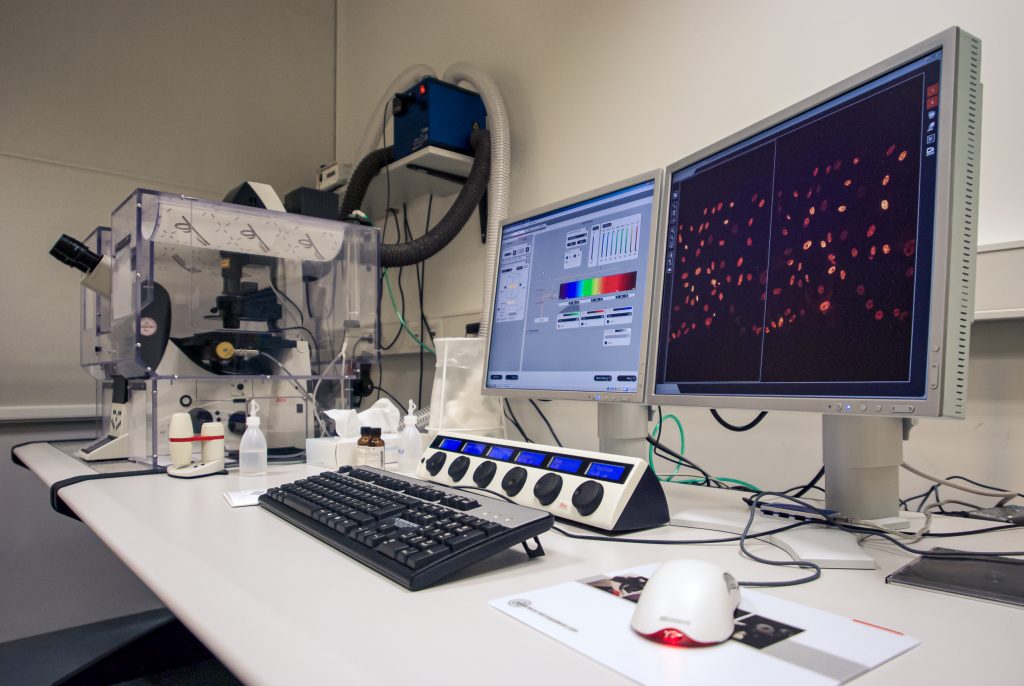
Commercial Leica TCS-SP5 setup in my facility at the TU Dresden; despite the heating enclosure, the microscope was mostly used for fixed samples 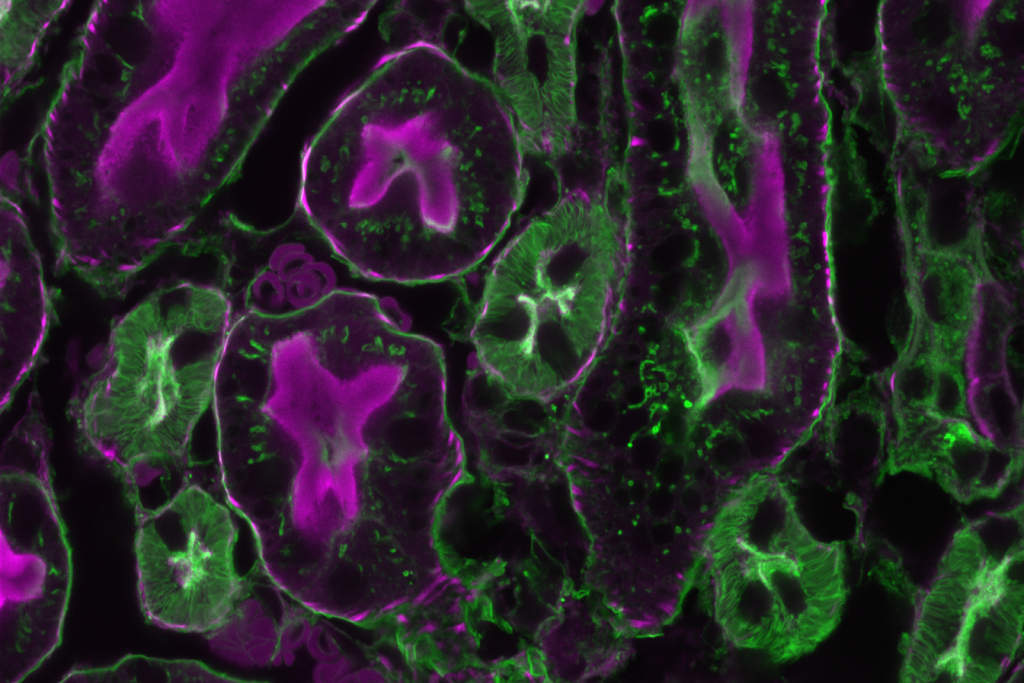
Image of a fixed mouse kidney section recorded on this microscope
My first exposure to light sheet microscopy was during a demo of an early Zeiss prototype of what eventually became the Lightsheet.Z1 (now Lightsheet 7) called ‘Demonstrator B’. Later, while working on my master thesis in the lab of Pavel Tomancak, I got the chance to work with the microscope to image fruit fly embryogenesis. The results coming from this compact little microscope were stunning to me, even though computers back in the days were struggling to save and process the stream of time-lapse image data. I had high hopes for such a microscope to become available commercially soon after. Unfortunately that only happened much later, and those light sheet setups were surprisingly expensive. On the upside, it inspired the microscopy community to come up with home-built light sheet microscopes like the OpenSPIM, a small and simple open-source setup.
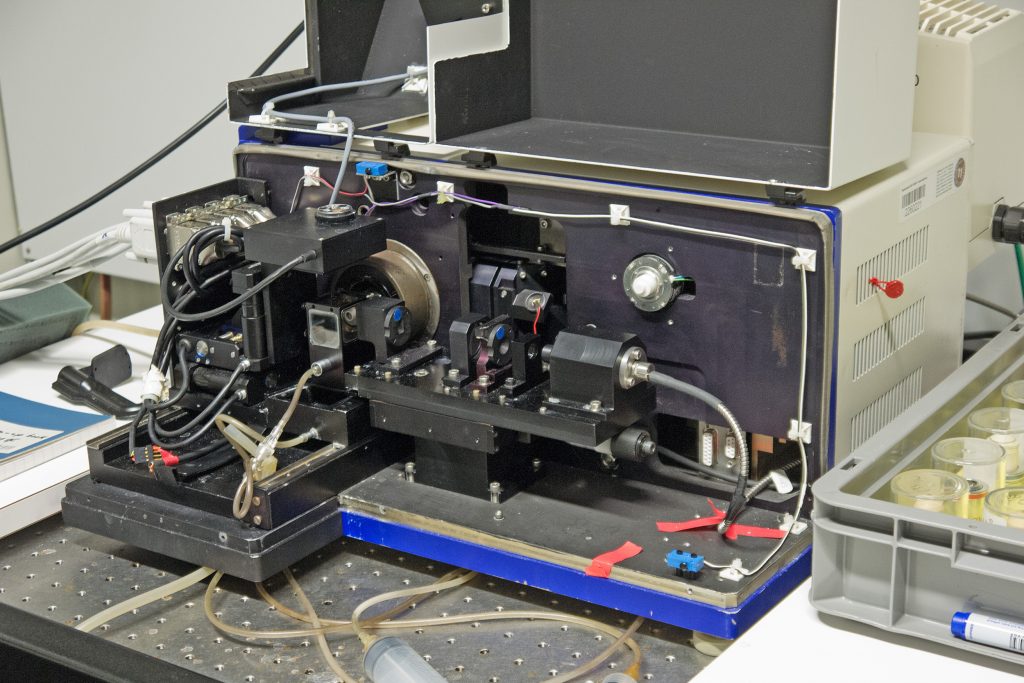
Early Zeiss ‘Demonstrator B’ light sheet microscope at the MPI-CBG 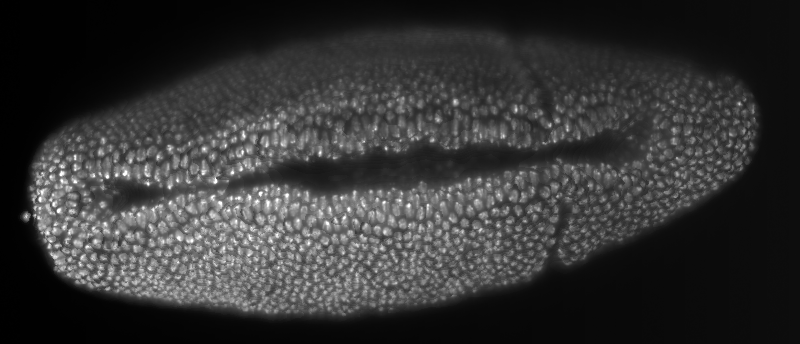
Smooth manifold extraction (SME) of a developing Drosophila melanogaster embryo with nuclei marker recorded with the Zeiss prototype 
Pavel Tomancak and Peter Pitrone showing off the OpenSPIM in a suitcase (openspim.org) 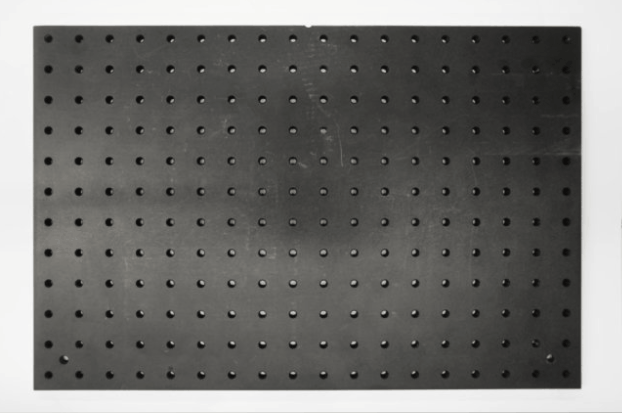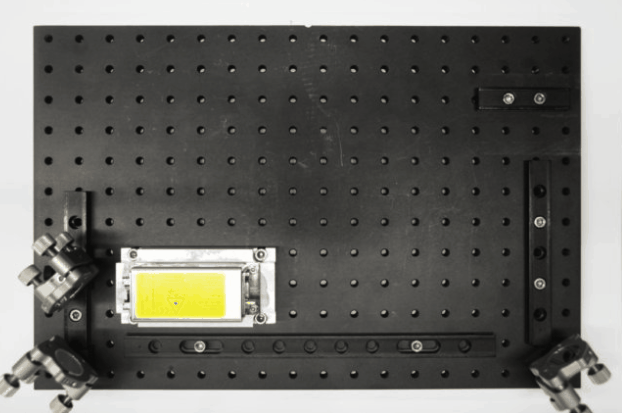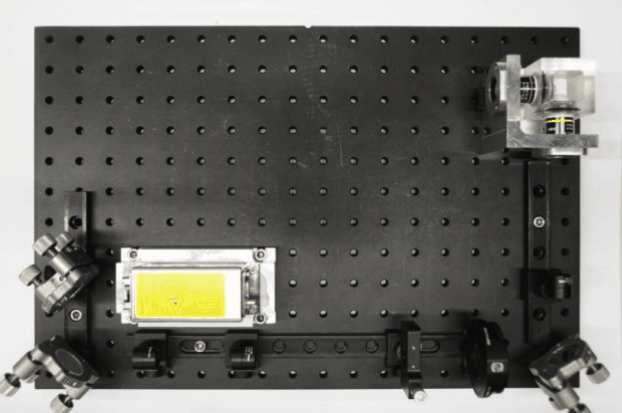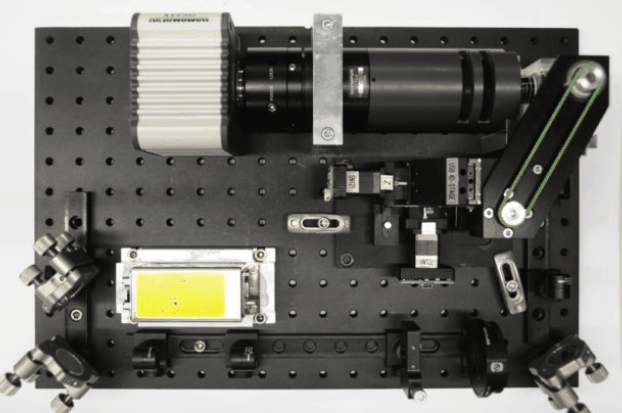
Time-lapse of an OpenSPIM being assembled (openspim.org)
Later on, when joining Jan Huisken’s lab at the MPI-CBG for my PhD, I realized that yes indeed, a custom-built microscope can deliver so much more insight into a biological sample. Jan co-invented light sheet microscopy in the early 2000s, and building custom setups using this technology in his lab showed me how to unlock the full potential of microscopes. Designing, building, testing and troubleshooting my own research-grade microscope for studying and manipulating the cardiovascular system in zebrafish turned out to be a long process. Selecting the right off-the-shelf parts, spending hours and hours designing useful and not-so useful parts in a 3D software, testing cameras and setting up a matching computer, filling shelfs with power supplies and controllers, connecting tons of cables and much more. In addition, a lot of time and effort went into the software, which was tailor-made for this microscope by Benjamin Schmid. In the end everything worked out great, and Michaela Mickoleit and I (with the help of Benjamin and Nico Scherf) got awesome 3D data of the zebrafish heart and its underlying function with the help of ‘SPIM3’. It didn’t work that well for some other samples, though, because everything from the field of view to the software features was optimized for the zebrafish heart. The microscope setup was also fairly complex and stationary, so collaborators would have to bring or ship their sample to us for using this light sheet microscope.
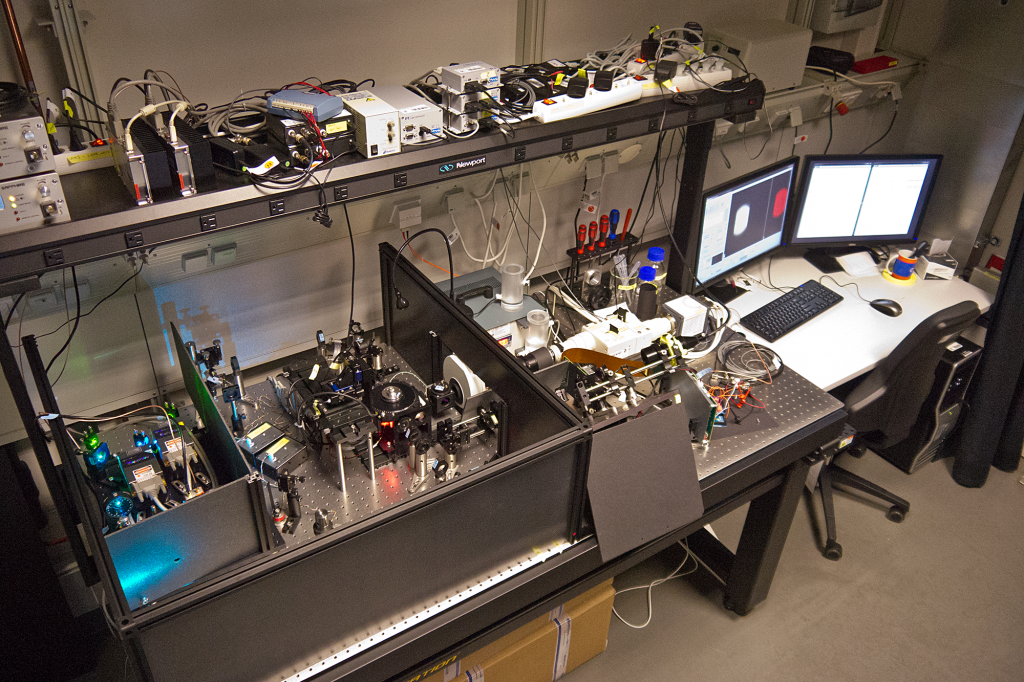
My SPIM3 setup in the Huisken lab at the MPI-CBG 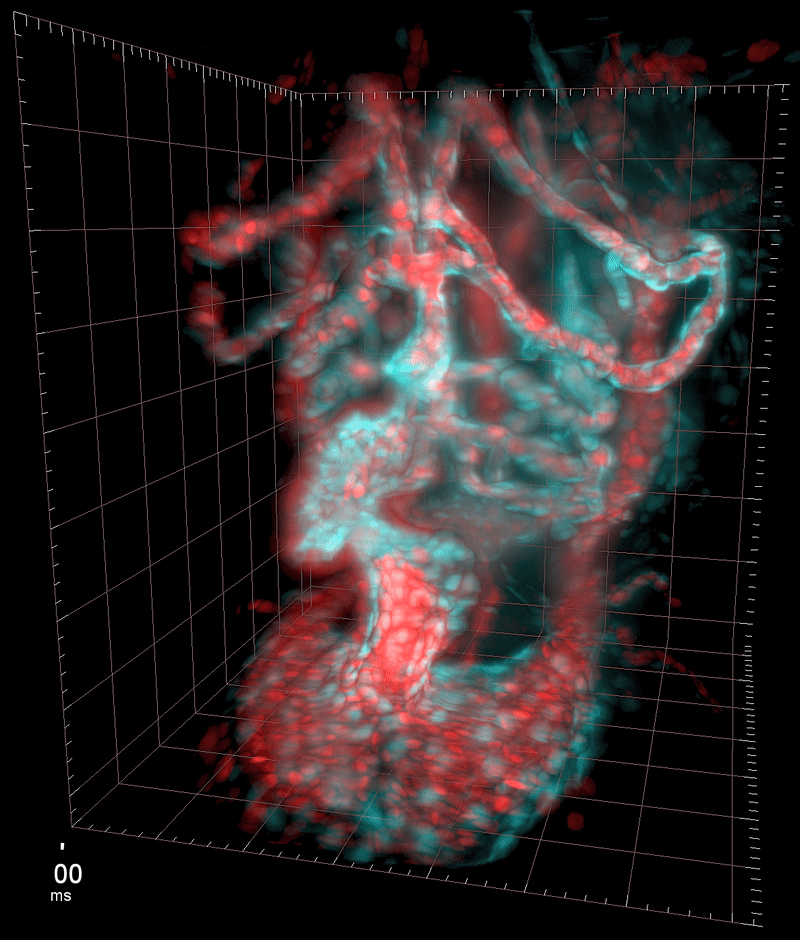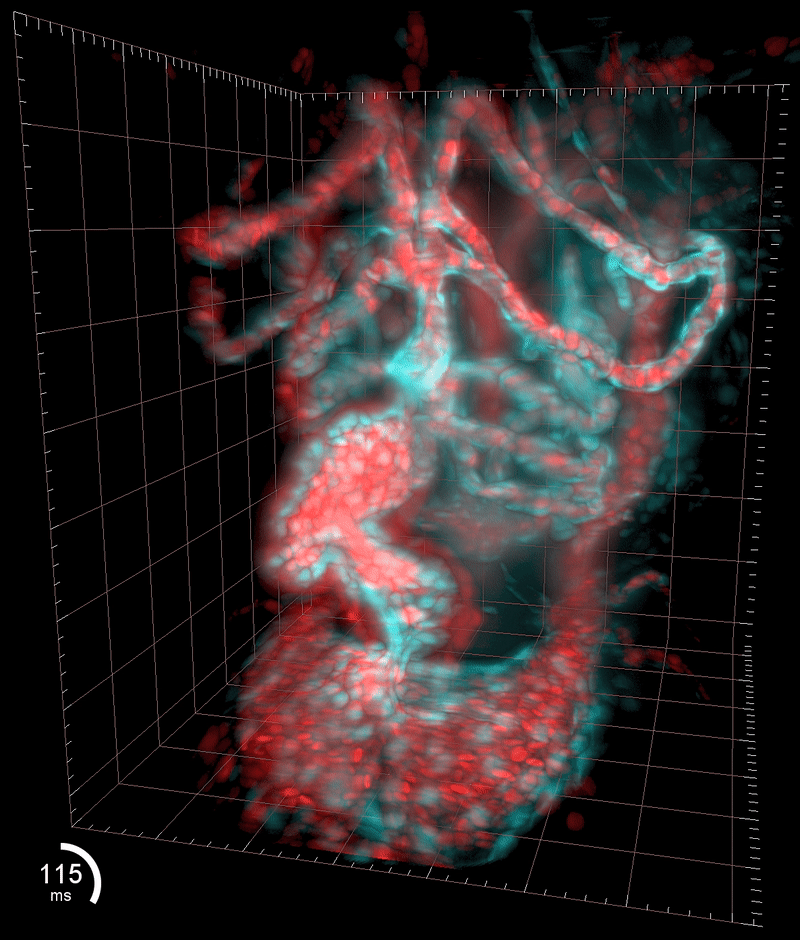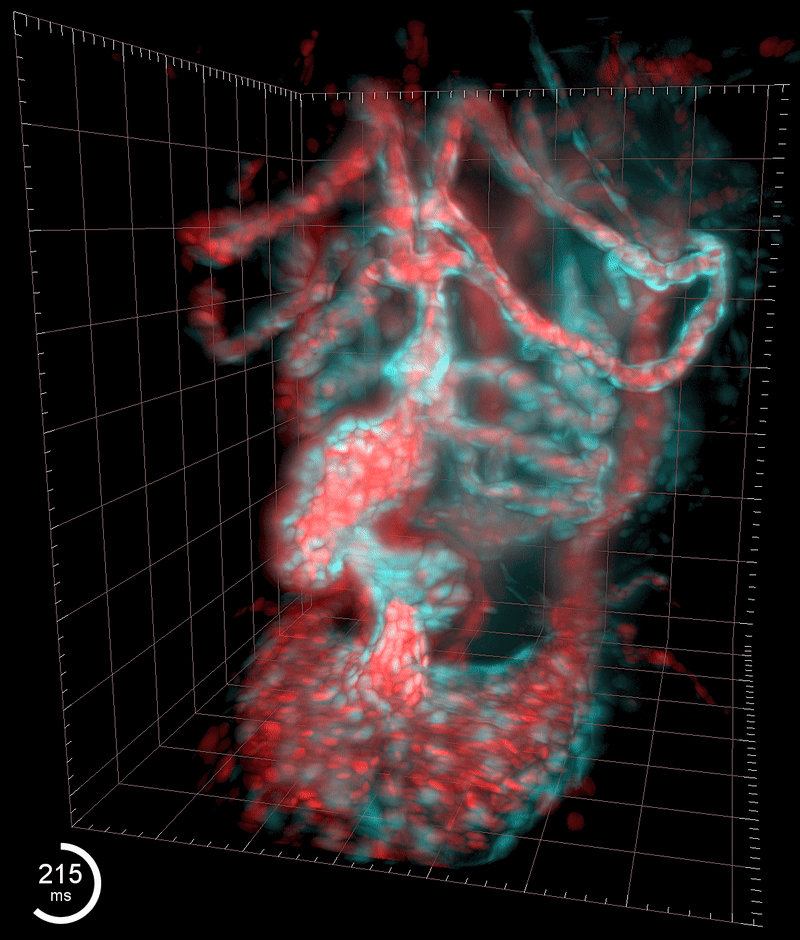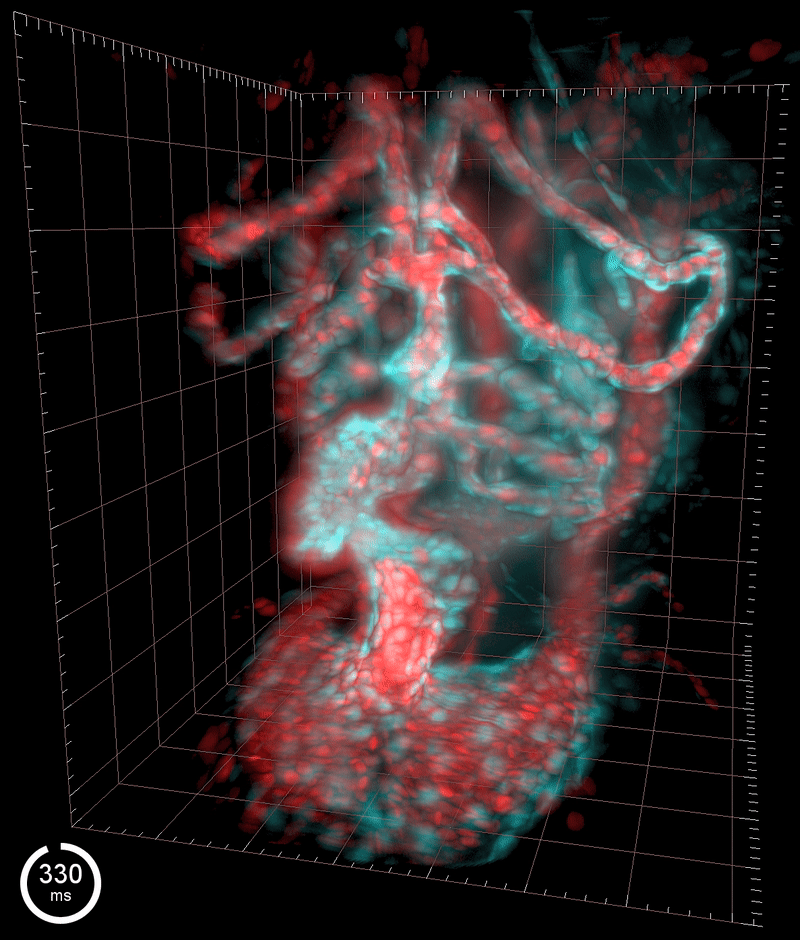
Three-dimensional reconstruction of the beating heart and surrounding vasculature in a 3-day-old zebrafish 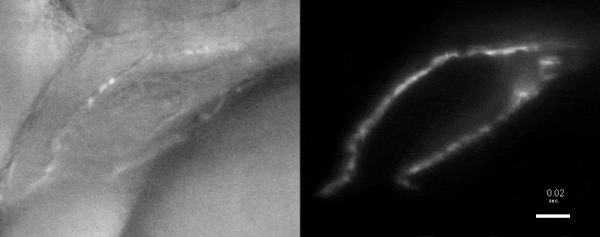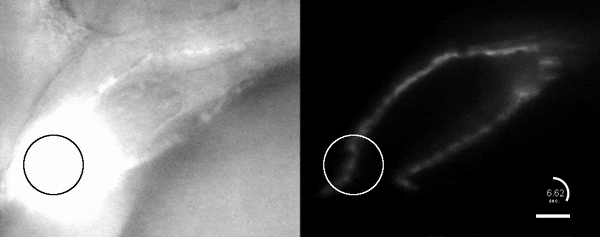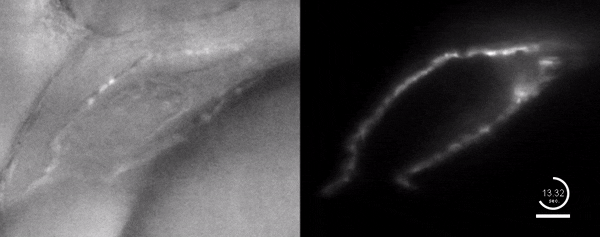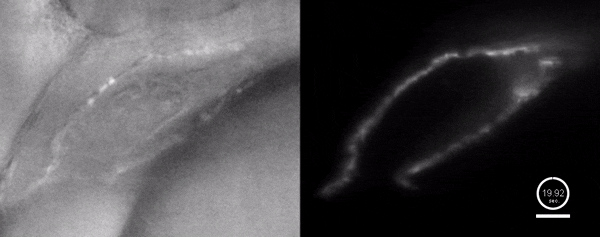
Optogenetic disruption of calcium flow in a 1-day-old zebrafish heart
When it was time for Jan to move on, SPIM3 had to be left behind. One of the reasons was that the microscope was just too complex, big and stationary to be moved across the ocean into the new lab. Instead, a second iteration was built with yet another software to run it. By that time, I had already left Jan’s lab for a postdoc position in Jennifer Waters’ Nikon Imaging Center (NIC) at Harvard Medical School (HMS) here in Boston, MA. Besides learning a ton about microscopy, teaching, and running a core facility, I also got the chance to build a light sheet microscope. We as a core facility needed it to be portable enough to come along for our annual light microscopy course in Cold Spring Harbor Laboratory (CSHL), but I also wanted it to be research-grade enough to be able to test light sheet microscopy with local scientists. I could have built an OpenSPIM, but decided to come up with my own. Designing and building my setup outside of an actual optics development lab – without all the knowledge and tools readily available – was both fun and challenging at the same time. Putting the software together in LabView took me some time, but eventually everything worked. My microscope turned out larger than I wanted in the beginning and it would just fit in a big car to be driven down to CSHL. It did perform well, though, and besides using it for workshops I worked with a couple of researchers from HMS and their samples. I would have had to move the microscope between buildings and get closer to the individual labs to cover more projects involving live samples, but the size and weight prevented me from doing that on a regular basis.

The light sheet microscope I built in the NIC, packed up in the trunk of a large car 
The setup in action during our annual light microscopy course at CSHL (photo by Talley Lambert) 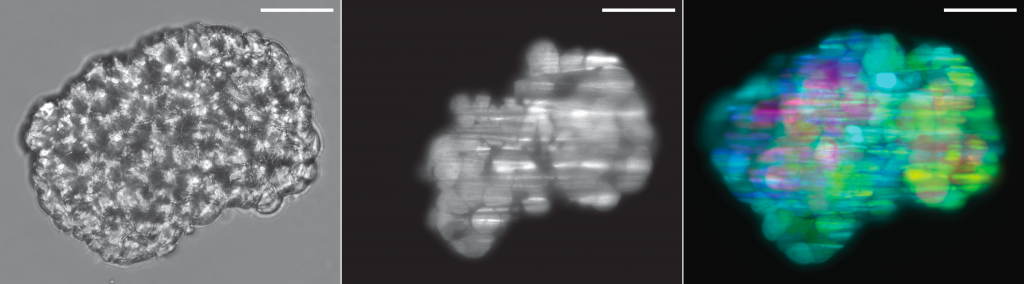
Cancer cell spheroid recorded on this system: brightfield, optical section and depth-projection (sample by Marcin Iwanicki)
While I was working in the NIC, Jan established his new lab at the Morgridge Institute for Research in Madison, WI. He and I kept in touch, and just when I was thinking about my next steps he told me about a new light sheet microscopy project he and members of his lab were working on. The idea was to design and build compact microscopes comprised of modules such that each system can quickly be customized for a specific application. The microscopes could then be packed and shipped to collaborators for a given time, before being returned for changes and updates. I was intrigued by what would be a radically new model of giving researchers access to the latest microscopy technology. Sometime later I re-joined Jan’s lab and took over the role of application specialist for that project. By then, the microscope was given the name ‘Flamingo’ and the team had already built an impressive prototype. Rory Power and Todd Bakken took the same optics used in big stationary microscopes, folded them up and used custom machined and 3D-printed parts to pack them up in a compact and portable unit. The integration of mSPIM eliminated most of the striping artifacts known from more basic light sheet microscopes, and which are also visible in the cancer spheroid recorded on my NIC setup above. The Flamingo prototype even had its own laser combiner and could be converted between the classic SPIM configuration with the sample oriented vertically and a tilted one with the specimen mounted in a dish just by exchanging a handful of parts. To be able to control the microscope, Joe Li developed electronics and software that adapt to whatever modules are installed. Due to its compactness, the Flamingo prototype could be placed on a bench, rolled from place to place on a cart, and packed in a case to take it on a plane to Germany.

Todd working on the first Flamingo prototype 
Joe working on custom electronics for the Flamingo 
Rory mounting the emission filter on a Flamingo microscope 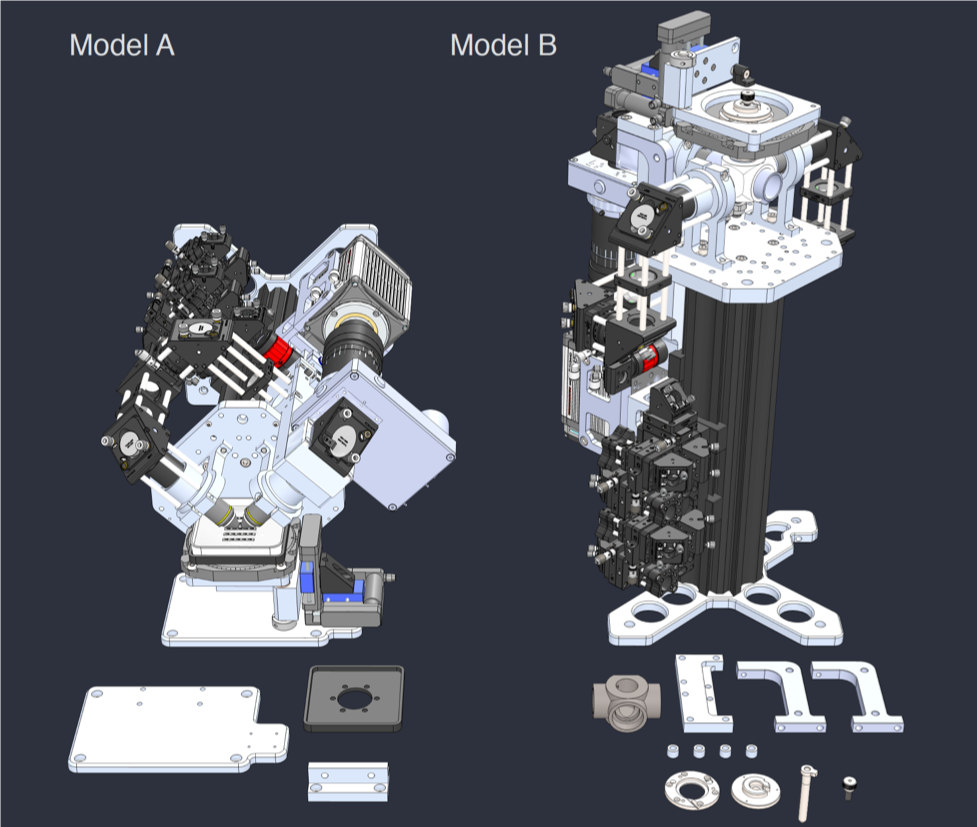
Rendering of the first Flamingo prototype and the parts required to convert it from one configuration to the other
While testing the first Flamingo prototype in our lab, we made a long list of things to tweak and came up with a new iteration. It turned out a lot smaller and way more polished, supports four different configurations and has a high level of modularity. The entire setup fits in two larger suitcases and can be moved from lab to lab, packed in the trunk of a car and shipped around the world. Setting it up only takes about an hour, and our software has remote-control options to supervise the microscope from afar. Now it was time to go out and test the Flamingo in other research labs, and this is what I am here for. The team in Madison sent me a Flamingo and I started collaborations with biologists in several institutions in the Boston area who always wanted to try light sheet microscopy. The Zon and Fishman labs at Harvard University kindly offered space for me to use as ‘home base’ where I can use the Flamingo on my own, run tests and invite other scientists to take a first look at the microscope and discuss project ideas. The Flamingo performs well with a variety of biological samples, including zebrafish, worms and plants, even in long time-lapses. The results are on par with images recorded on our well-proven, but large and stationary microscopes. I trained many scientists on advanced light microscopes, and the Flamingo sessions stand out as being closer to the actual research, because we are working in the scientist’s ‘personal space’ close to where the actual sample resides. In return, we are collecting great feedback about the capabilities and usability of hard- and software, which helps us to decide what we need to change for the next iteration of the Flamingo.

A Flamingo microscope in a fish room at Harvard University 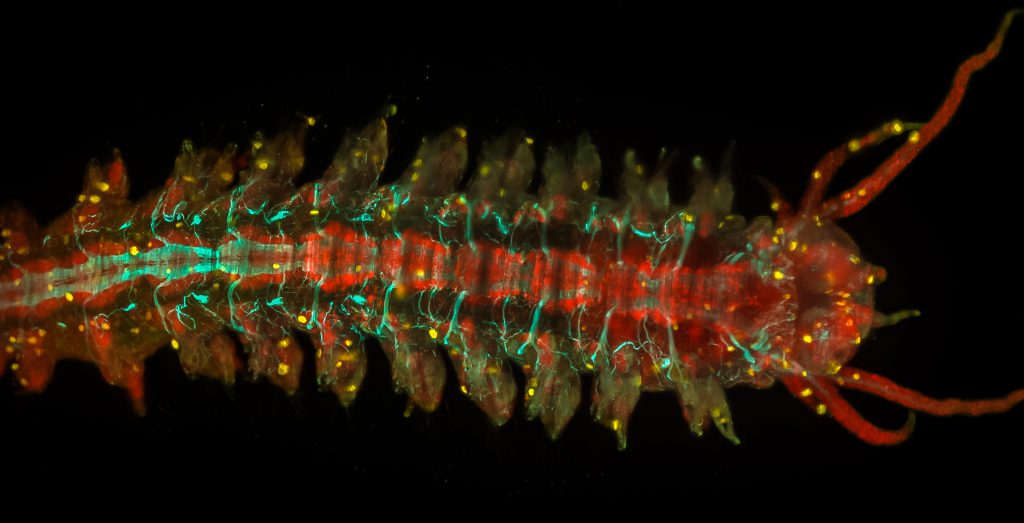
Adult Platynereis dumerilii (sample by Duygu Özpolat) 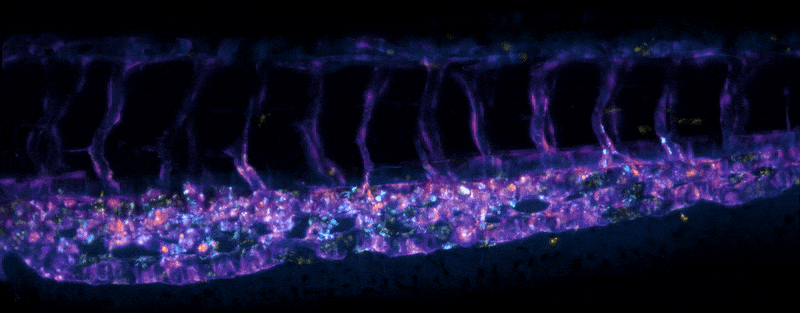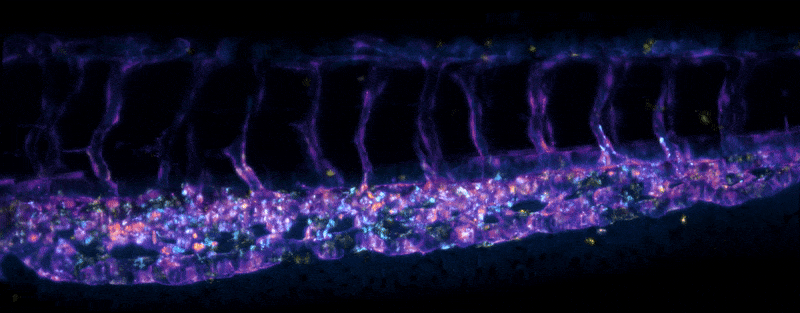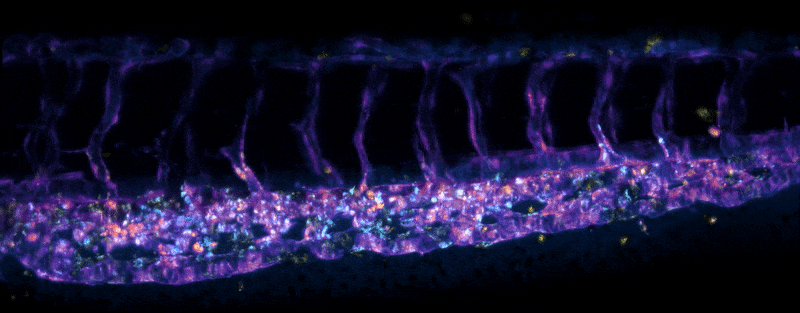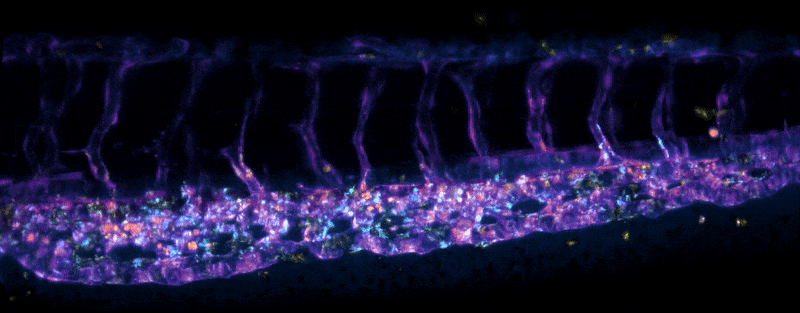
Stem cells migrating in the tail of a larval zebrafish (sample by Elliot Hagedorn) 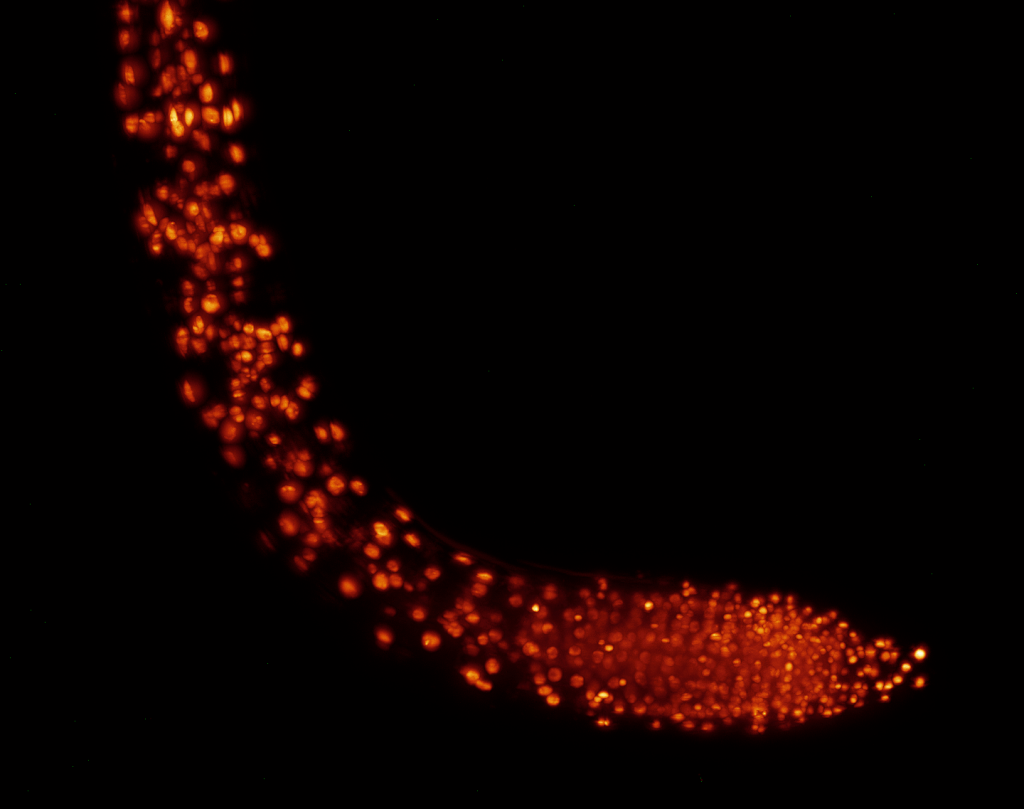
Growth zone of an Arabidopsis thaliana root (sample by Cara Winter) 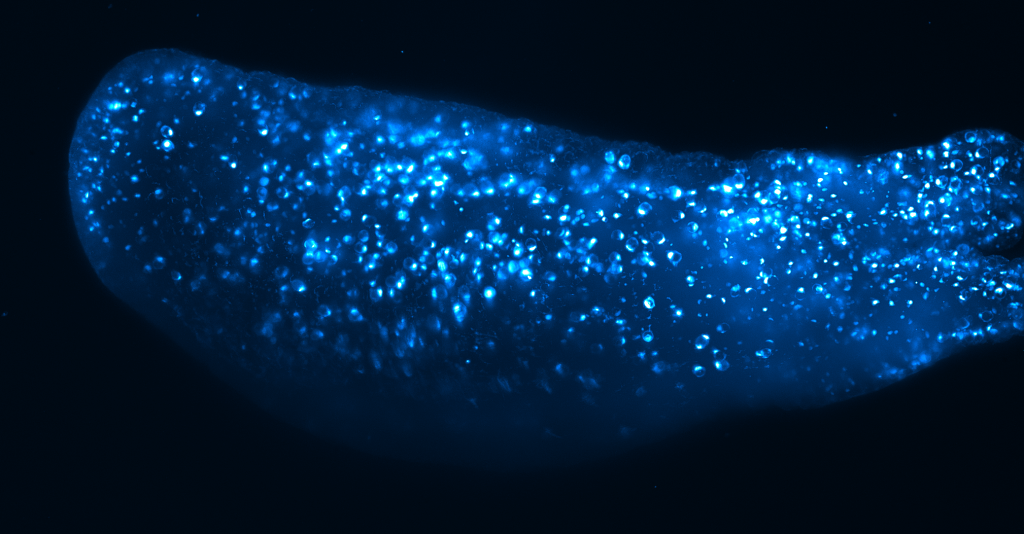
Hydra (sample by Alison Hanson) 
Me working on a Flamingo microscope at Boston Children’s Hospital
Besides research collaborations, we also use the Flamingo for workshops. There, having more of an open system is great for giving attendees a real sense of how things work. Unfortunately, we will have to wait before attending the next hands-on workshop, and several of our collaborations are delayed due to the pandemic. In times where people cannot get together and sharing became much harder, the Flamingo does have some advantages over traditional microscopes, though. Running experiments requires very little hands-on with the microscope itself, and both microscope control and data storage are decoupled and completely in the hands of each individual user. In addition, the Flamingo control system allows to connect remotely, which means we no longer have to be in the same room (or even building/city/country) to use the microscope together with a collaborator. And being so small, we can place a Flamingo on the desk next to us and use it for online demos or during virtual talks and workshops.
Jan at one of his light sheet microscopes (mpi-cbg.de) 
A Flamingo microscope in Jan’s home office (photo by Jan Huisken)
With Flamingo, we revolutionize how and where high-powered research microscopes are setup and used. Our project has transformative benefits for biologists, as it allows them to do studies on living organisms close to where they reside. With help from our supporters, we aim to democratize high-end light microscopy, bringing it to campuses and labs for free. We implemented this concept using light sheet microscopy, because that is what we know best. Of course, we would love to see the idea spreading through the microscopy community. We see large potential in expanding our concept to areas beyond light sheet microscopy, and we would be happy to get new partners on board to push this new approach forward. If you are interested in using a Flamingo or contributing to our project, find out more at involv3d.org.


 (6 votes, average: 1.00 out of 1)
(6 votes, average: 1.00 out of 1)