A career path to bioimage analysis
Posted by Laurent Thomas, on 9 December 2020
I am currently working in Heidelberg, Germany, finishing my PhD thesis between the medical university of Heidelberg and the microscopy company ACQUIFER. My research project is dedicated to the development of user-oriented software solutions (Fiji plugins, KNIME workflows…) to facilitate the handling and analysis of large microscopy datasets of 2D images. The project is motivated by screening studies in zebrafish, but I tried to develop tools that are broadly applicable. The project is part of the Horizon2020/MSCA ITN ImageInLife [1], which funded 14 PhD student positions over the last 3 years in different research institutions, including industrial partners such as ACQUIFER.
I am now working as a so-called bioimage analyst/research-software engineer/data-scientist. I got to this career path progressively. I first studied physics, chemistry and maths back in my home-town, preparing a highly selective exam to enter an engineering school (specific to the French education system and similar to a master program). After 2 hard-working years, I finally entered the biotechnology school of Strasbourg (ESBS). At that time, I did not have any biology lecture since high-school, but I managed to catch up with the theory. We also had a lot of wet-lab hands-on, which were completely new to me, but lots of fun although I did all kind of beginner’s mistakes. Back there I also had my first introduction to python programming, I did not really have time to dedicate to it but from time to time I was writing some toy scripts or following some online tutorial, which in the end helped me when I started using python for image-processing.
Along with the training in Strasbourg, we also had a few weeks of hands-on and lectures in Freiburg, Germany and at the Biozentrum in Basel, Switzerland, covering topics from plant physiology to microbiology. Again, very interesting experience and great time with the classmates. I also did an exchange-semester in the BIOTEC master program in Dresden. At a conference there I met Benoit Lombardot, who was working in the image-analysis facility in the MPI-CBG, together with Robert Haase. We got on well and I came back a few months later for a 1-month internship at the facility. Although it was just 1 month, I believe it was a key to my career path. That’s when I started getting serious about image-analysis! During this month, I started using KNIME and python, working on a 3D cell tracking pipeline. By chance, Christian Dietz from KNIME was around for a week of conference at the MPI, so I also got to know him, and he showed me some tricks to improve the workflow!
I finished the 3-year biotech engineer program in Strasbourg with a 6-month internship in the start-up Abbelight in Paris, working on hardware interfacing and a bit of image-processing for super-resolution microscopy. The company was just about a year old, so I was actually their first trainee! About halfway through the internship, I heard about the PhD program ImageInLife via the university in Strasbourg. The position in Heidelberg was really matching my skills and interests so I applied, and I got the job! I finished the internship at Abbelight in August 2017, I graduated in Strasbourg end of September and I started right away the PhD in October. Fun fact, during the first year I had the chance to attend some conferences (MIFOBIO, Neubias, Quantitative Bioimaging) where I was regularly seeing my former colleagues from Abbelight!
I have learned a lot about open-source, image-analysis and research-software engineering during this PhD project. This was possible thanks to community resources such as the ImageJ wiki, but also thanks to active support from other bioimage analysts and developers on the forum (image.sc). I also have a very supportive supervisor at ACQUIFER, Jochen Gehrig, whose feedback and guidance helped me developing user-friendly software tools. I try my best to make accessible and well documented tools, because I know that software documentation is often a limiting factor for bioimage analysis. For some contributions, we actually published software papers, but besides the article I usually provide extensive documentation in the GitHub repositories, as well as video tutorials on my YouTube channel or on the official Fiji YouTube channel. However, I must say it represents a major effort to make this documentation, and you don’t get much “academic” recognition for it, as it often does not even count for an article. I believe this lack of recognition is unfortunately common to research-software engineers and bioimage analysts, but I hope it will change in the future with the spread of open-science practices.
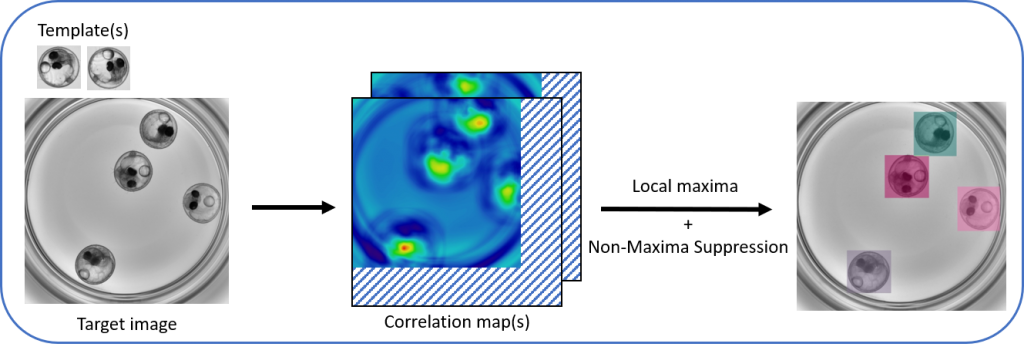
A first outcome of my PhD was multi-template-matching, a method for object-detection by bounding-boxes, which is more robust than single-template-matching and simpler to use than e.g. machine-learning object-detectors. We use it to localize specific organs or tissues in our images of zebrafishes, but it can work with a wide range of objects. I implemented the method as Fiji plugins and as a python package, so you are quite flexible to use it in custom workflows.
I also worked with other object-detection and image-classification methods, and I found that there was a lack of solutions to quickly annotate regions-of-interests (ROI) and categories in ImageJ/Fiji. At some point, I became comfortable enough with the ImageJ java source code, so I proposed the implementation of a category attribute associated to ROIs, to allow the annotation of different object-categories in an image (see the pdf in the Neubias F1000 gateway and this forum post). I was super proud when Wayne Rasband, the original author of ImageJ, accepted my proposal and implemented the changes I suggested!
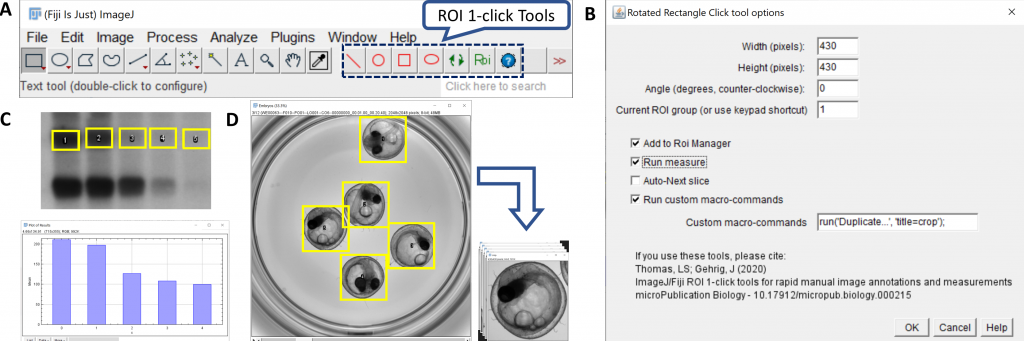
Following that, I also developed some toolsets to facilitate routine image-annotations for large number of ROIs or images, first with the ROI 1-click tools (which are contained in a single ImageJ macro script by the way !), and lately with plugins for the annotations of images or ROIs with multiple keywords (qualitative annotation plugins).
One major challenge for me is to spread the word about these contributions. I try to bring a poster every time I go to a conference, and I even had the chance to give a flash talk at the NEUBIAS 2019 conference in Luxembourg. Still, I found it challenging to reach end-users who are not involved in the bioimage analysis community. Alternatively, besides documenting the project, YouTube and GitHub help reaching a broader audience, eventhough the satisfaction is not really the same… I also use open-science repositories, such as Zenodo, on which we uploaded example datasets, posters and archived versions of software tools. I really like the communities functionality, so I can directly associate an upload to the Neubias, ImageInLife and even the ACQUIFER community space all at once. I also reference my tools on the BioImage Informatics Index (biii.eu) or on the KNIME Hub. I think those resources have a great potential but are still largely unexplored. I definitely encourage bioimage analysts and microscopists to check them out and to raise awareness about them. I am also moderately active on Twitter (@LauLauThom) and on the image.sc forum, which are handy to monitor new contributions and software functionalities (I advise watching the announcement section of the forum and @Talk_BioImg on twitter). Following my PhD, I have the chance to continue working with ACQUIFER as a full-time employee, starting next year, to work on internal software and hardware development. I will likely have less time to dedicate to open-source community contributions, although I will still work with my favourite open-source software from time to time, and I still have some resources from my PhD to explore 😉

Note
[1] This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 721537 “ImageInLife”



 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)