Tissue clearing: what invisible samples reveal about biology
Posted by jorgealmagr, on 8 September 2021
Written by Jorge Almagro and Hendrik Messal
The tissues that constitute organs exist in our body in 3D. However, for practical reasons, histological analysis has been traditionally performed in 2D, slicing a few micrometer-thick sections to analyse them under the microscope. While this has been critical to understand our anatomy for centuries, it constitutes a simplification of our complex 3D architecture. In the past decade, an effort has been made to visualise tissues with cellular resolution in 3D. Due to the pigmentation and opacity of most mammalian tissues, the light penetration and scattering in whole mounts or thick sections has traditionally limited 3D imaging. To overcome this problem, several techniques have been developed to clarify tissues. Once tissues become transparent, it is possible to see through them, and with genetically encoded fluorophores, antibody staining or dyes, visualise the 3D organisation of molecules, cells, and structures.
Tissue clearing techniques
Over 60 clearing techniques have been published in peer review journals in the last decade. While the original clearing techniques were developed to image the nervous system, other organs have different physicochemical characteristics and require specific treatments for permeabilization, labelling and achieving transparency. Hence, bespoke techniques have been developed to image specific organs, and some laboratories have made efforts to image heterogeneous samples such as whole organisms successfully.
Clearing techniques are usually classified based on the physicochemical strategy used to label and clear tissues: 1. Electrophoresis-mediated, 2. Organic solvents. 3. Aqueous solutions. Samples are usually permeabilised with detergents or alcohols, labelled as required, and cleared by immersion in a refractive index-matching solution. Each technique may be optimal to clear a specific specimen, and detailed reviews listing these techniques are available1. The chemicals used in some of these techniques are toxic and challenging to handle in large volumes for immersion of whole organs and imaging. Other considerations on chemical reagents used, such as compatibility with the tissue or molecule of interest are important to have in mind when choosing a clearing technique. In our latest review, we have provided a summary of chemical reagents used in permeabilization and clearing by refractive index matching with the hope that it can help researchers develop combinations for organs in which clearing has not yet been achieved2.
FLASH: our personal story
In our group, Axel Behrens’ laboratory at The Francis Crick Institute, we investigate the function of adult stem cells in epithelial organs (most of those containing hollow structures, like ducts and glands), and the cancers that originate in them, which we call carcinomas. These epithelial organs are generally stiffer and more difficult to clear than the brain, so we struggled to apply brain-specific clearing techniques to visualise the mouse pancreas and study its architecture in physiology and disease. While some of the available techniques would allow us to detect the hormone-producing islets of Langerhans and the digestive enzyme-secreting acinar cells, they failed to retrieve specific markers of the ductal network that transports the pancreatic juices to the small intestine. These ducts are key in the study of carcinoma, as they constitute the cell of origin of pancreatic cancer. To be able to visualise the pancreatic ducts, we performed a high-throughput comparison of detergents at different times and temperatures3. We discovered that a simple incubation overnight in SDS-borate solution at 54oC was optimal to visualise the entire ductal network of a mouse pancreas. This is the basis of FLASH (Fast Light-microscopic Analysis of antibody Stained-wHole organs). In further experiments we discovered that these initial FLASH conditions, ideal to image the pancreas, did not retrieve certain epitopes in dense organs like the liver, and could destroy fragile samples such as E13.5 or earlier mouse embryos. Hence, in a Gedankenexperiment we hypothesised zwitterionic detergents (polar but with a net charge of 0) would permeabilise dense organs better, and will also protect vulnerable samples in comparison to the harsh polarity of a charged detergent like SDS. Indeed, Zwittergent 3-10 in combination with urea and borate proved to be the ideal recipe to clear mouse livers and embryos.
Currently we have several modifications and specific procedures to apply FLASH to very diverse samples, including mouse epithelial organs, other highly pigmented organs like the heart and the spleen, embryos at different stages of development, human samples, and entirely different organisms like Drosophila melanogaster.
Imaging cleared tissue
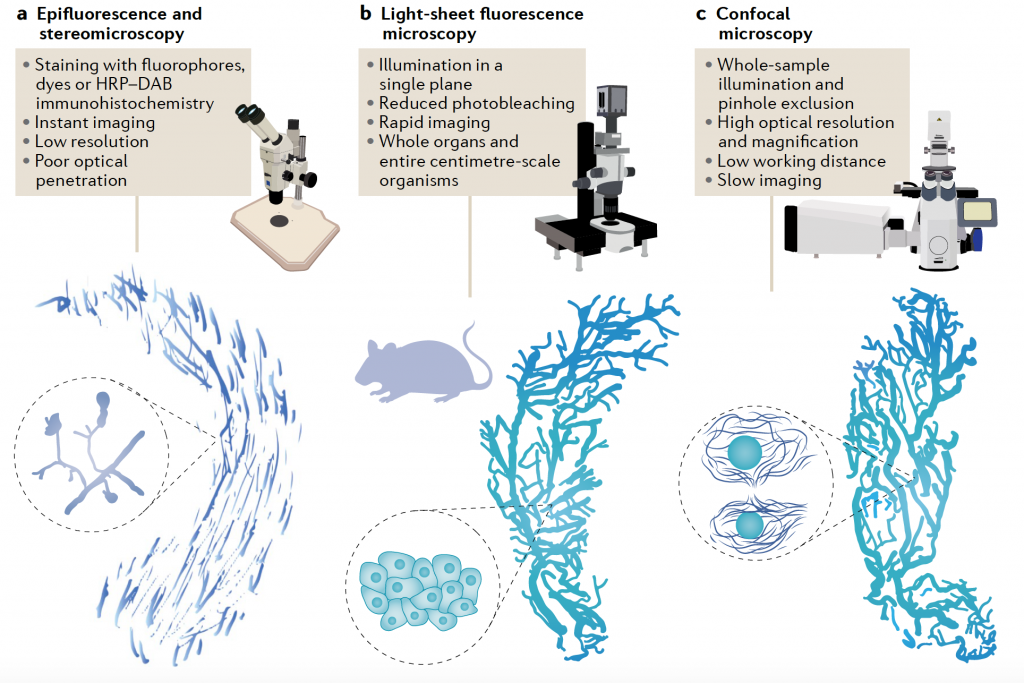
Microscopes to image cleared tissues might also differ depending on the desired resolution and magnification. While it is possible to take simple images of cleared mouse organs (or samples of similar size) in stereomicroscopes and epifluorescence microscopes, most research articles utilise light-sheet fluorescence (LSFM) or confocal microscopes (CM). LSFM is the preferred technique for imaging increasingly large, cleared samples. While subcellular resolution is rarely achieved in this system, it allows for rapid image of entire organs and organisms. New microscopy developments have allowed to even image whole pig and human organs4. CM is preferrable for high-resolution imaging, but slower to image large samples. However, improvements in CM such as spinning disk allow the high-resolution imaging of entire mouse organs in a matter of hours.
Clearing tumours exposes their complexity
Tissue clearing started as an approach taken by neuroscientists to better visualise the complexity of the brain (in fact they revisited the idea from anatomist Werner Spalteholtz, who cleared organs more than a century ago). Since the combination of clearing and fluorescence imaging was successfully achieved, tissue clearing has expanded to many different types of samples and, as a consequence, different fields in biology. Specifically in the field of cancer, tissue clearing has great potential in basic research, as well as in clinical diagnostics. Tumours can be extremely intrinsically heterogeneous in structures, patterns, and cellular composition. High-resolution 3D imaging of entire preclinical tumours or patient resections brings important advantages: on one hand, more data can be extracted per sample than with conventional histology, in addition, rare events (niches, cells…) that may be missed with 2D histology are easier to catch by imaging entire specimens, and finally, details on the structural architecture of a tumour, which can be informative of cancer behaviour, prognosis or response to therapy, can be grasped.
Using tissue clearing and mouse models of pancreatic cancer, we were able to identify the 3D growth pattern of early lesions, inwards or outwards the duct of origin (which we named endophytic and exophytic lesions respectively). This differential growth was predictive of prognosis, with exophytic lesions being able to recruit cancer associated fibroblasts that enhanced tumour growth5. Other researchers have used tissue clearing to study tumorigenesis in complex multi-layered epithelia6, to study metastatic spread unbiasedly in entire organisms7, and to characterise the tumour microenvironment and its role in tumour biology, among other applications.
Researchers are also making an effort to bring clearing and 3D imaging to cancer diagnostics. 3D clearing of patient material could bring great advantages to improve the power and accuracy of histopathology. For example, assessing lymph node metastasis using entire cleared samples could significantly reduce misdiagnosis. In addition, architectural aspects of tumours or vasculature only observable by clearing and 3D imaging may have valuable prognostic value and be used for patient stratification.
In conclusion, tissue clearing is a thriving method that brings high resolution fluorescence imaging thick tissue sections, whole organs, and even entire organisms, revolutionising the way we look at biological samples.
Authors

Jorge Almagro
PhD Student
The Francis Crick Institute

Dr. Hendrik Messal
Postdoctoral Fellow
Netherlands Cancer Institute (NKI)
References
1 Richardson, D. S. & Lichtman, J. W. Clarifying Tissue Clearing. Cell 162, 246-257, doi:10.1016/j.cell.2015.06.067 (2015).
2 Almagro, J., Messal, H. A., Zaw Thin, M., van Rheenen, J. & Behrens, A. Tissue clearing to examine tumour complexity in three dimensions. Nat Rev Cancer, doi:10.1038/s41568-021-00382-w (2021).
3 Messal, H. A. et al. Antigen retrieval and clearing for whole-organ immunofluorescence by FLASH. Nat Protoc 16, 239-262, doi:10.1038/s41596-020-00414-z (2021).
4 Zhao, S. et al. Cellular and Molecular Probing of Intact Human Organs. Cell 180, 796-812 e719, doi:10.1016/j.cell.2020.01.030 (2020).
5 Messal, H. A. et al. Tissue curvature and apicobasal mechanical tension imbalance instruct cancer morphogenesis. Nature 566, 126-130, doi:10.1038/s41586-019-0891-2 (2019).
6 Fiore, V. F. et al. Mechanics of a multilayer epithelium instruct tumour architecture and function. Nature585, 433-439, doi:10.1038/s41586-020-2695-9 (2020).
7 Pan, C. et al. Deep Learning Reveals Cancer Metastasis and Therapeutic Antibody Targeting in the Entire Body. Cell 179, 1661-1676 e1619, doi:10.1016/j.cell.2019.11.013 (2019).


 (4 votes, average: 1.00 out of 1)
(4 votes, average: 1.00 out of 1)