Microscopy Open Access collection
Posted by FocalPlane, on 25 October 2021
This week is International Open Access Week 2021 (25-31 October). To celebrate it, we have created a collection of Open Access articles on microscopy from the five journals of The Company of Biologists: Development, Journal of Cell Science, Journal of Experimental Biology, Disease Models & Mechanisms and Biology Open.
We hope you enjoy our microscopy collection:
PRIMERS
Tissue clearing and 3D imaging in developmental biology. Alba Vieites-Prado, Nicolas Renier. Development (2021) 148 (18): dev199369.
REVIEWS
Deep learning for bioimage analysis in developmental biology. Adrien Hallou, Hannah G. Yevick, Bianca Dumitrascu, Virginie Uhlmann. Development (2021) 148 (18): dev199616.
Of numbers and movement – understanding transcription factor pathogenesis by advanced microscopy. Julia M. T. Auer, Jack J. Stoddart, Ioannis Christodoulou, Ana Lima, Kassiani Skouloudaki, Hildegard N. Hall, Vladana Vukojević, Dimitrios K. Papadopoulos. Dis Model Mech (2020) 13 (12): dmm046516.
RESEARCH ARTICLES AND REPORTS
Live imaging of the Drosophila ovarian niche shows spectrosome and centrosome dynamics during asymmetric germline stem cell division. Gema Villa-Fombuena, María Lobo-Pecellín, Miriam Marín-Menguiano, Patricia Rojas-Ríos, Acaimo González-Reyes. Development (2021) 148 (18): dev199716.
Live imaging of adult zebrafish cardiomyocyte proliferation ex vivo. Hessel Honkoop, Phong D. Nguyen, Veronique E. M. van der Velden, Katharina F. Sonnen, Jeroen Bakkers. Development (2021) 148 (18): dev199740.
Anterior expansion and posterior addition to the notochord mechanically coordinate zebrafish embryo axis elongation. Susannah B. P. McLaren, Benjamin J. Steventon. Development (2021) 148 (18): dev199459.
The Arabidopsis stomatal polarity protein BASL mediates distinct processes before and after cell division to coordinate cell size and fate asymmetries. Yan Gong, Julien Alassimone, Andrew Muroyama, Gabriel Amador, Rachel Varnau, Ao Liu, Dominique C. Bergmann. Development (2021) 148 (18): dev199919.
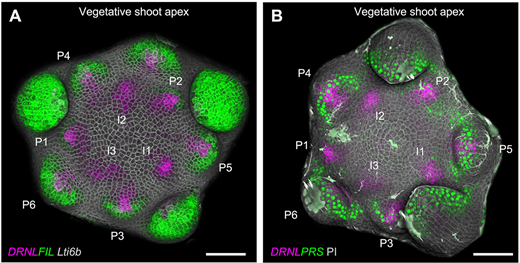
Stable establishment of organ polarity occurs several plastochrons before primordium outgrowth in Arabidopsis. Feng Zhao, Jan Traas. Development (2021) 148 (18): dev198820.
Collective nuclear behavior shapes bilateral nuclear symmetry for subsequent left-right asymmetric morphogenesis in Drosophila. Dongsun Shin, Mitsutoshi Nakamura, Yoshitaka Morishita, Mototsugu Eiraku, Tomoko Yamakawa, Takeshi Sasamura, Masakazu Akiyama, Mikiko Inaki, Kenji Matsuno. Development (2021) 148 (18): dev198507.
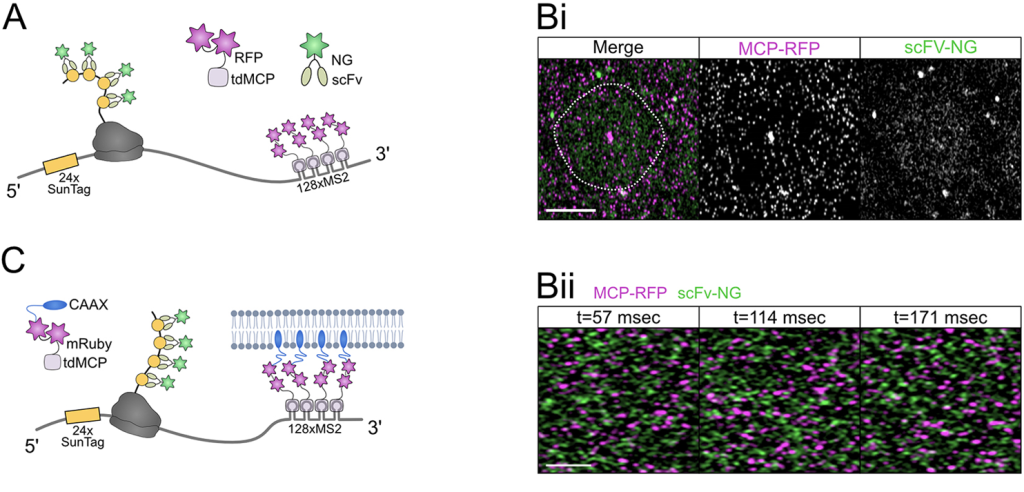
Dynamics of hunchback translation in real-time and at single-mRNA resolution in the Drosophila embryo. Daisy J. Vinter, Caroline Hoppe, Thomas G. Minchington, Catherine Sutcliffe, Hilary L. Ashe. Development (2021) 148 (18): dev196121.
Structural variability and dynamics in the ectodomain of an ancestral-type classical cadherin revealed by AFM imaging. Shigetaka Nishiguchi, Hiroki Oda. J Cell Sci (2021) 134 (14): jcs258388.
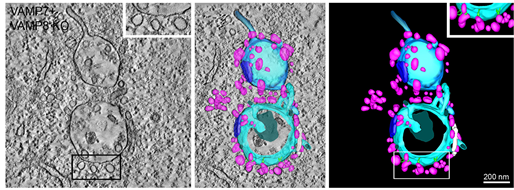
Organelle tethering, pore formation and SNARE compensation in the late endocytic pathway. Luther J. Davis, Nicholas A. Bright, James R. Edgar, Michael D. J. Parkinson, Lena Wartosch, Judith Mantell, Andrew A. Peden, J. Paul Luzio. J Cell Sci (2021) 134 (10): jcs255463.
Hijacking of the host cell Golgi by Plasmodium berghei liver stage parasites. Mariana De Niz, Reto Caldelari, Gesine Kaiser, Benoit Zuber, Won Do Heo, Volker T. Heussler, Carolina Agop-Nersesian. J Cell Sci (2021) 134 (10): jcs252213.
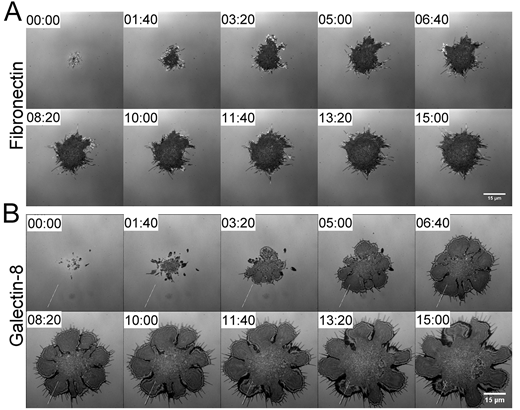
Differential cellular responses to adhesive interactions with galectin-8- and fibronectin-coated substrates. Wenhong Li, Ana Sancho, Wen-Lu Chung, Yaron Vinik, Jürgen Groll, Yehiel Zick, Ohad Medalia, Alexander D. Bershadsky, Benjamin Geiger. J Cell Sci (2021) 134 (8): jcs252221.
The formin inhibitor SMIFH2 inhibits members of the myosin superfamily. Yukako Nishimura, Shidong Shi, Fang Zhang, Rong Liu, Yasuharu Takagi, Alexander D. Bershadsky, Virgile Viasnoff, James R. Sellers. J Cell Sci (2021) 134 (8): jcs253708.
Brain endothelial tricellular junctions as novel sites for T cell diapedesis across the blood–brain barrier. Mariana Castro Dias, Adolfo Odriozola Quesada, Sasha Soldati, Fabio Bösch, Isabelle Gruber, Tobias Hildbrand, Derya Sönmez, Tejas Khire, Guillaume Witz, James L. McGrath, Jörg Piontek, Masuo Kondoh, Urban Deutsch, Benoît Zuber, Britta Engelhardt. J Cell Sci (2021) 134 (8): jcs253880.
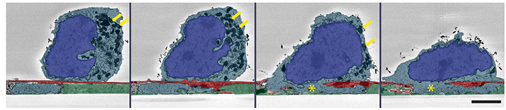
Natural killer cell immune synapse formation and cytotoxicity are controlled by tension of the target interface. Daniel Friedman, Poppy Simmonds, Alexander Hale, Leoma Bere, Nigel W. Hodson, Michael R. H. White, Daniel M. Davis. J Cell Sci (2021) 134 (7): jcs258570.
L-selectin regulates human neutrophil transendothelial migration. Izajur Rahman, Aida Collado Sánchez, Jessica Davies, Karolina Rzeniewicz, Sarah Abukscem, Justin Joachim, Hannah L. Hoskins Green, David Killock, Maria Jesus Sanz, Guillaume Charras, Maddy Parsons, Aleksandar Ivetic. J Cell Sci (2021) 134 (3): jcs250340.
Hydrostatic pressure prevents chondrocyte differentiation through heterochromatin remodeling. Koichiro Maki, Michele M. Nava, Clémentine Villeneuve, Minki Chang, Katsuko S. Furukawa, Takashi Ushida, Sara A. Wickström. J Cell Sci (2021) 134 (2): jcs247643.
Interpreting the pathogenicity of Joubert syndrome missense variants in Caenorhabditis elegans. Karen I. Lange, Sofia Tsiropoulou, Katarzyna Kucharska, Oliver E. Blacque. Dis Model Mech (2021) 14 (1): dmm046631.
Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms. Ottilie von Loeffelholz, Andrew Purkiss, Luyan Cao, Svend Kjaer, Naoko Kogata, Guillaume Romet-Lemonne, Michael Way, Carolyn A. Moores. Biol Open (2020) 9 (7): bio054304.
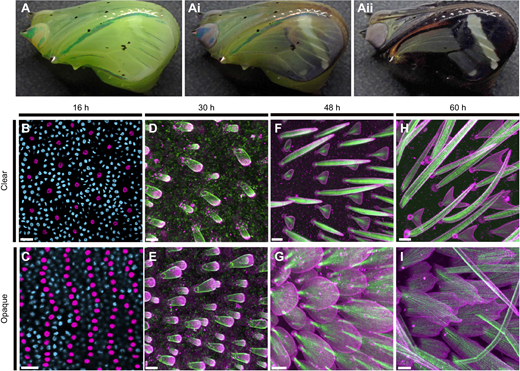
Developmental, cellular and biochemical basis of transparency in clearwing butterflies. Aaron F. Pomerantz, Radwanul H. Siddique, Elizabeth I. Cash, Yuriko Kishi, Charline Pinna, Kasia Hammar, Doris Gomez, Marianne Elias, Nipam H. Patel. J Exp Biol (2021) 224 (10): jeb237917.
The wing scales of the mother-of-pearl butterfly, Protogoniomorpha parhassus, are thin film reflectors causing strong iridescence and polarization. Doekele G. Stavenga. J Exp Biol (2021) 224 (15): jeb242983.
TECHNIQUES AND RESOURCES
QuantifyPolarity, a new tool-kit for measuring planar polarized protein distributions and cell properties in developing tissues. Su Ee Tan, Weijie Tan, Katherine H. Fisher, David Strutt. Development (2021) 148 (18): dev198952.
MOrgAna: accessible quantitative analysis of organoids with machine learning. Nicola Gritti, Jia Le Lim, Kerim Anlaş, Mallica Pandya, Germaine Aalderink, Guillermo Martínez-Ara, Vikas Trivedi. Development (2021) 148 (18): dev199611.
NaNuTrap: a technique for in vivo cell nucleus labelling using nanobodies.
Zsuzsa Ákos,Leslie Dunipace,Angelike Stathopoulos. Development (2021) 148 (18): dev199822.
Microfluidic-based imaging of complete Caenorhabditis elegans larval development. Simon Berger, Silvan Spiri, Andrew deMello, Alex Hajnal. Development (2021) 148 (18): dev199674.
LAMA: automated image analysis for the developmental phenotyping of mouse embryos. Neil R. Horner, Shanmugasundaram Venkataraman, Chris Armit, Ramón Casero, James M. Brown, Michael D. Wong, Matthijs C. van Eede, R. Mark Henkelman, Sara Johnson, Lydia Teboul, Sara Wells, Steve D. Brown, Henrik Westerberg, Ann-Marie Mallon. Development (2021) 148 (18): dev192955.
Python-Microscope – a new open-source Python library for the control of microscopes. David Miguel Susano Pinto, Mick A. Phillips, Nicholas Hall, Julio Mateos-Langerak, Danail Stoychev, Tiago Susano Pinto, Martin J. Booth, Ilan Davis, Ian M. Dobbie. J Cell Sci (2021) 134 (19): jcs258955.
Visualizing endogenous Rho activity with an improved localization-based, genetically encoded biosensor. Eike K. Mahlandt, Janine J. G. Arts, Werner J. van der Meer, Franka H. van der Linden, Simon Tol, Jaap D. van Buul, Theodorus W. J. Gadella, Joachim Goedhart. J Cell Sci (2021) 134 (17): jcs258823.
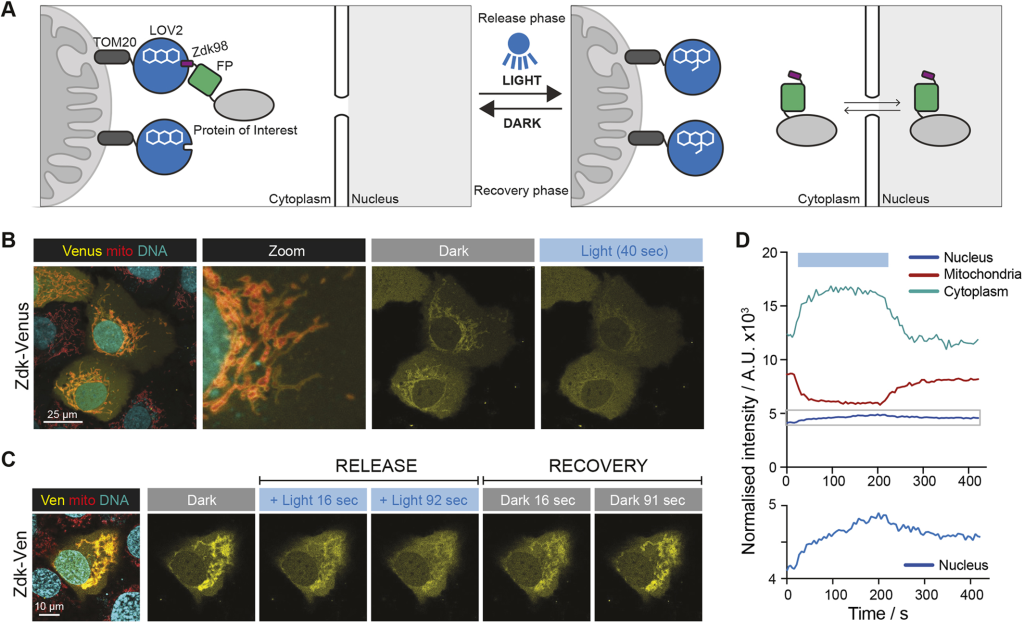
An optogenetic method for interrogating YAP1 and TAZ nuclear–cytoplasmic shuttling. Anna M. Dowbaj, Robert P. Jenkins, Daniel Williamson, John M. Heddleston, Alessandro Ciccarelli, Todd Fallesen, Klaus M. Hahn, Reuben D. O’Dea, John R. King, Marco Montagner, Erik Sahai. J Cell Sci (2021) 134 (13): jcs253484.
Direct observation of aggregate-triggered selective autophagy in human cells. Anne F. J. Janssen, Giel Korsten, Wilco Nijenhuis, Eugene A. Katrukha, Lukas C. Kapitein. J Cell Sci (2021) 134 (19): jcs258824.
Inducible manipulation of motor–cargo interaction using engineered kinesin motors. Jessica J. A. Hummel, Casper C. Hoogenraad. J Cell Sci (2021) 134 (15): jcs258776.
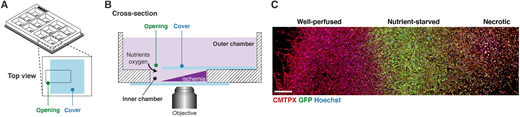
The MEMIC is an ex vivo system to model the complexity of the tumor microenvironment. Libuše Janská, Libi Anandi, Nell C. Kirchberger, Zoran S. Marinkovic, Logan T. Schachtner, Gizem Guzelsoy, Carlos Carmona-Fontaine. Dis Model Mech (2021) 14 (8): dmm048942.
Monitoring Schizosaccharomyces pombe genome stress by visualizing end-binding protein Ku. Chance E. Jones, Susan L. Forsburg. Biol Open (2021) 10 (2): bio054346.
Design of a 3D printed, motorized, uniaxial cell stretcher for microscopic and biochemical analysis of mechanotransduction. Noor A. Al-Maslamani, Abdulghani A. Khilan, Henning F. Horn. Biol Open (2021) 10 (2): bio057778.
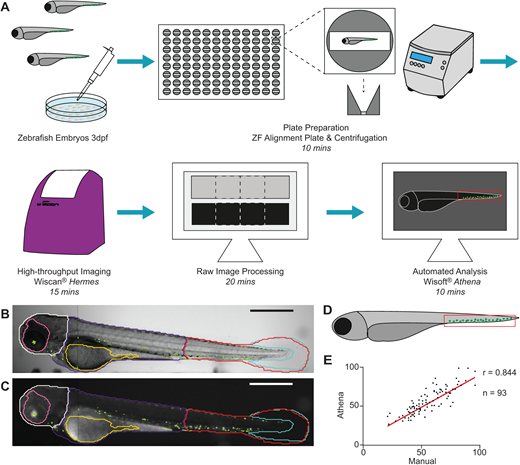
A versatile, automated and high-throughput drug screening platform for zebrafish embryos. Alexandra Lubin, Jason Otterstrom, Yvette Hoade, Ivana Bjedov, Eleanor Stead, Matthew Whelan, Gaia Gestri, Yael Paran, Elspeth Payne. Biol Open (2021) 10 (9): bio058513.
Quantitative analysis of subcellular distributions with an open-source, object-based tool. Pearl V. Ryder, Dorothy A. Lerit. Biol Open (2020) 9 (10): bio055228.
Read also our 2020 Open Access microscopy collection.


 (No Ratings Yet)
(No Ratings Yet)