Microscopy preprints – Bioimage analysis tools
Posted by FocalPlane, on 5 November 2021
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools only.
2020 BioImage Analysis Survey: Community experiences and needs for the future. Nasim Jamali, Ellen TA Dobson, Kevin W. Eliceiri, Anne E. Carpenter, Beth A. Cimini
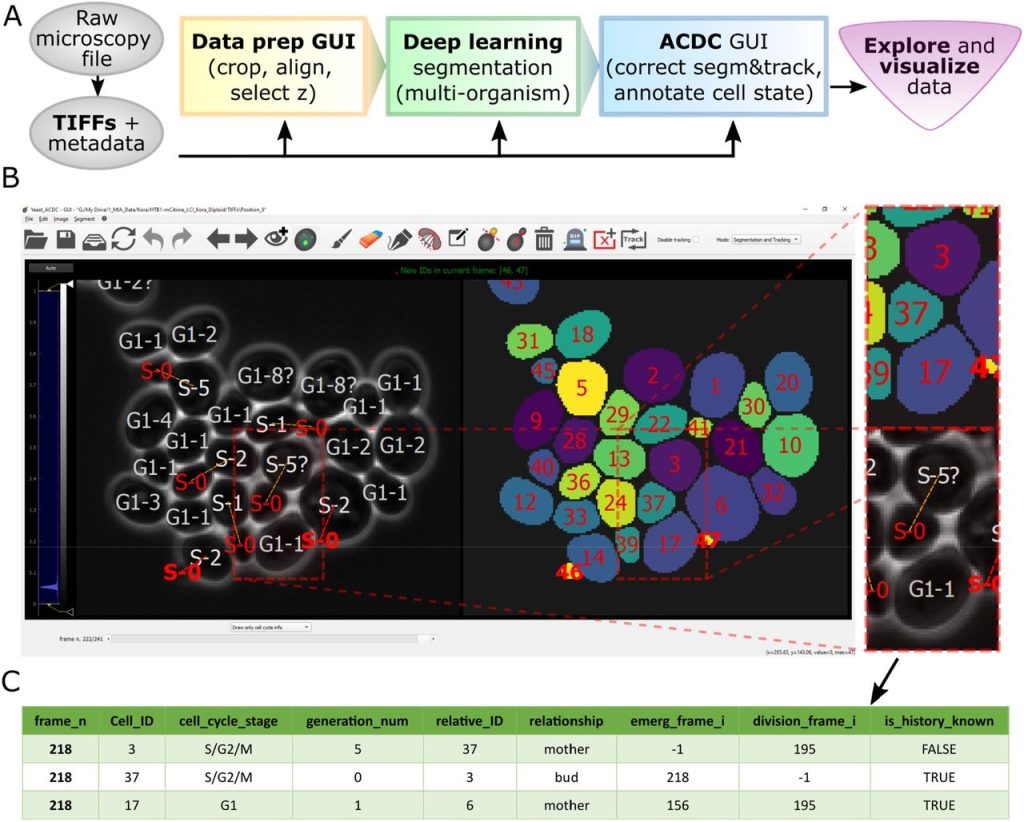
Cell-ACDC: a user-friendly toolset embedding state-of-the-art neural networks for segmentation, tracking and cell cycle annotations of live-cell imaging data. Francesco Padovani, Benedikt Mairhörmann, Pascal Falter-Braun, Jette Lengefeld, Kurt M. Schmoller
ZELDA: a 3D Image Segmentation and Parent-Child relation plugin for microscopy image analysis in napari. Rocco D’Antuono, Giuseppina Pisignano
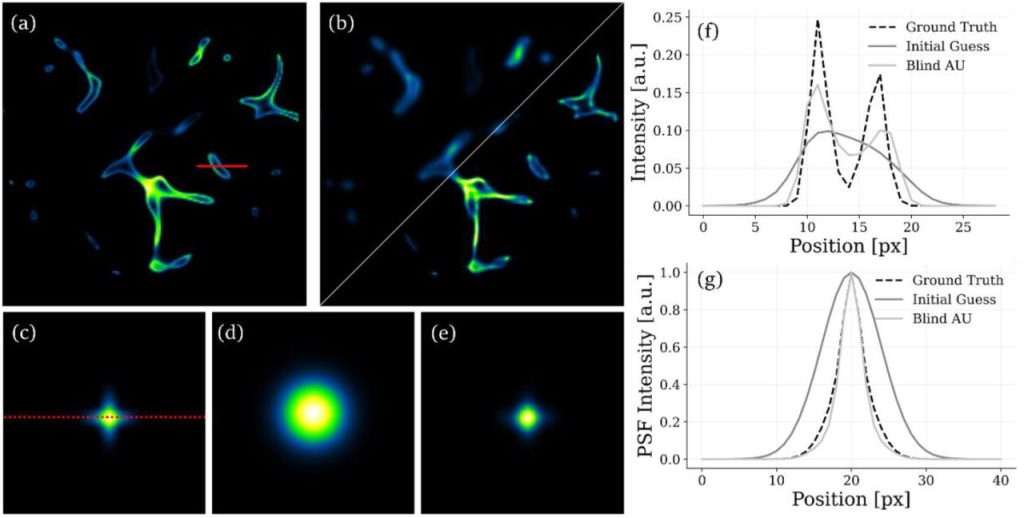
Blind deconvolution in autocorrelation inversion for multi-view light sheet microscopy. Elena Corbetta, Alessia Candeo, Andrea Bassi, Daniele Ancora
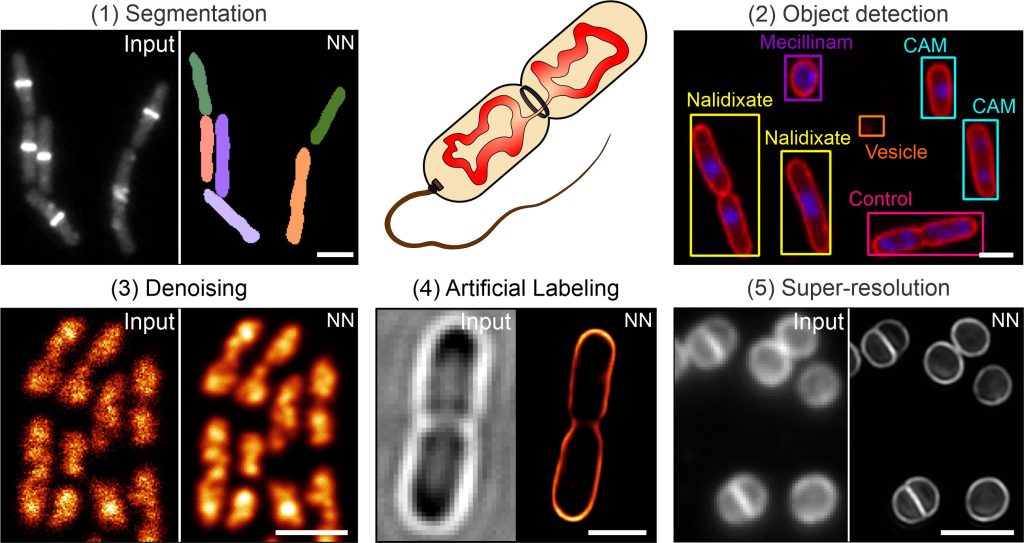
DeepBacs: Bacterial image analysis using open-source deep learning approaches. Christoph Spahn, Romain F. Laine, Pedro Matos Pereira, Estibaliz Gómez-de-Mariscal, Lucas von Chamier, Mia Conduit, Mariana G Pinho, Séamus Holden, Guillaume Jacquemet, Mike Heilemann, Ricardo Henriques
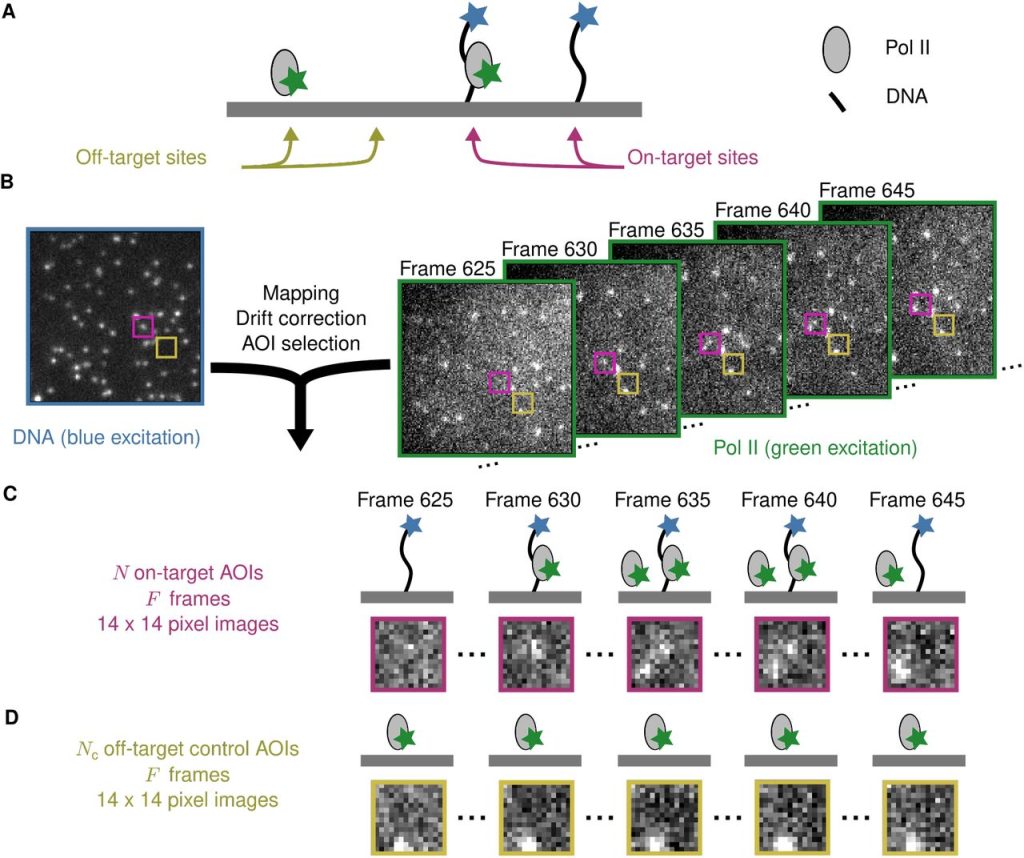
Bayesian machine learning analysis of single-molecule fluorescence colocalization images. Yerdos A. Ordabayev, Larry J. Friedman, Jeff Gelles, Douglas L. Theobald
DetecDiv, a deep-learning platform for automated cell division tracking and replicative lifespan analysis. Théo Aspert, Didier Hentsch, Gilles Charvin
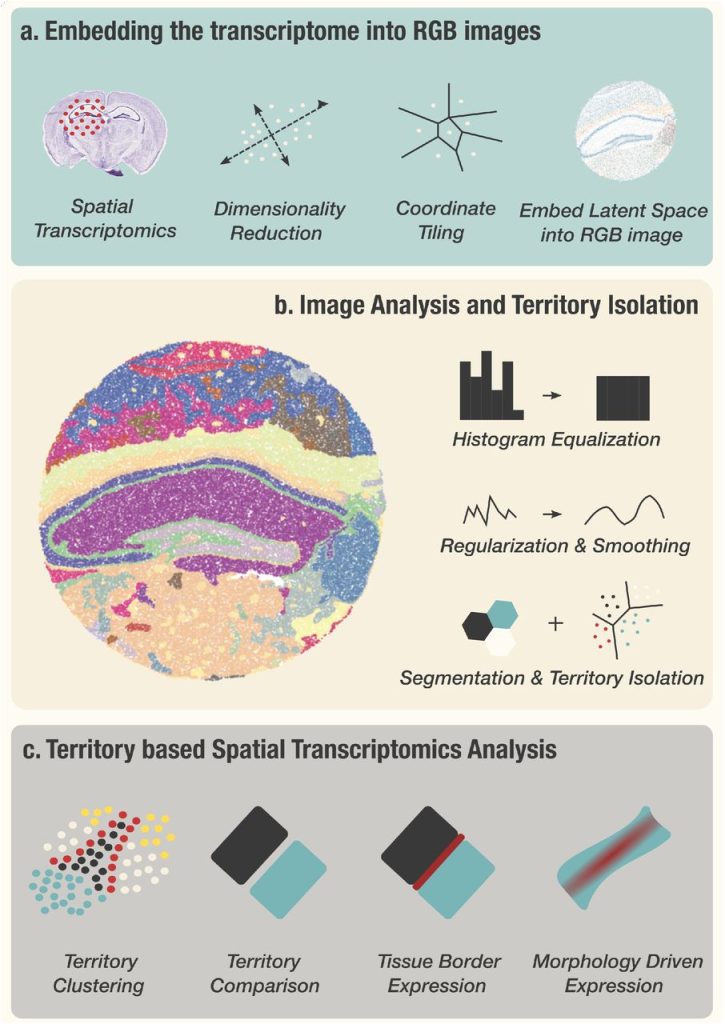
Vesalius: high-resolution in silico anatomization of Spatial Transcriptomic data using Image Analysis. Patrick C.N. Martin, Cecilia Lövkvist, Byung-Woo Hong, Kyoung Jae Won
An open-source method for analysis of confocal calcium imaging with sparse cells. Alisa A. Omelchenko, Lina Ni
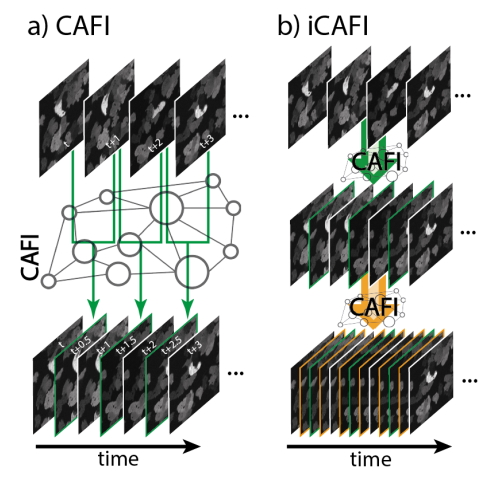
Content-aware frame interpolation (CAFI): Deep Learning-based temporal super-resolution for fast bioimaging. Martin Priessner, David C.A. Gaboriau, Arlo Sheridan, Tchern Lenn, Jonathan R. Chubb, Uri Manor, Ramon Vilar Compte, Romain F. Laine
YeastMate: Neural network-assisted segmentation of mating and budding events in S. cerevisiae. David Bunk, Julian Moriasy, Felix Thoma, Christopher Jakubke, Christof Osman, David Hörl
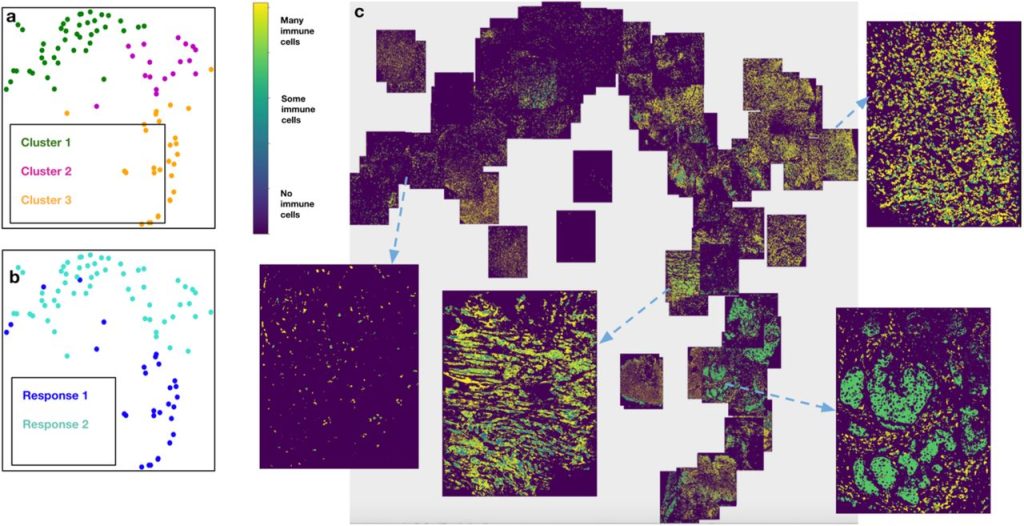
Mistic: an open-source multiplexed image t-SNE viewer. Sandhya Prabhakaran, Chandler Gatenbee, Mark Robertson-Tessi, Jeffrey West, Amer A. Beg, Jhanelle Gray, Scott Antonia, Robert A. Gatenby, Alexander R. A. Anderson
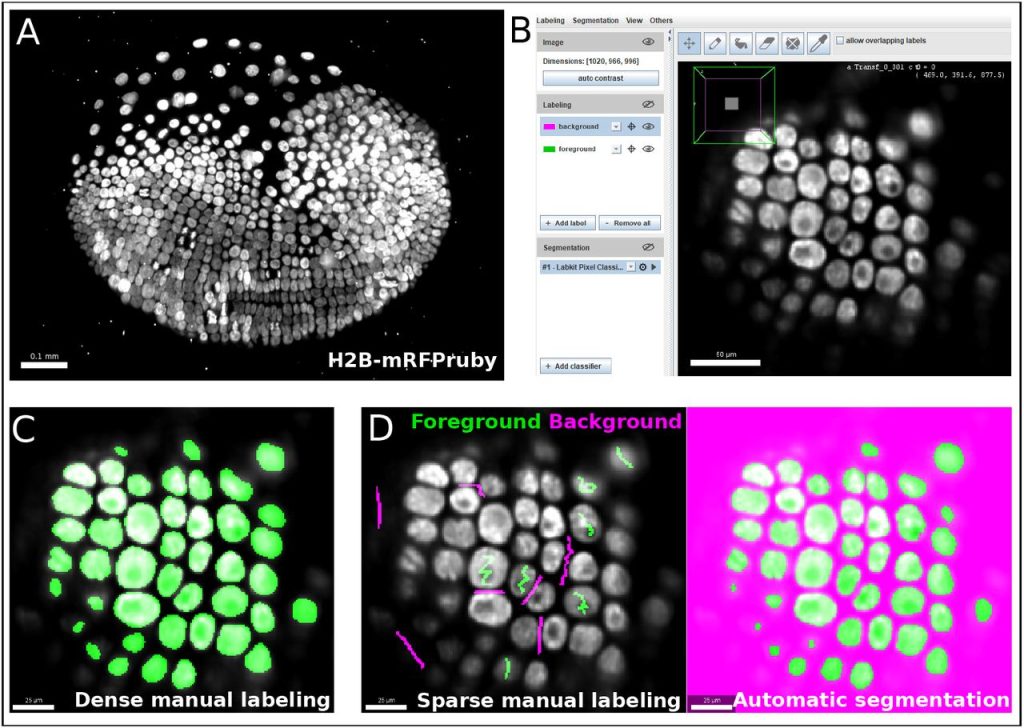
Labkit: Labeling and Segmentation Toolkit for Big Image Data. Matthias Arzt, Joran Deschamps, Christopher Schmied, Tobias Pietzsch, Deborah Schmidt, Robert Haase, Florian Jug
PySOFI: an open source Python package for SOFI. Yuting Miao, Shimon Weiss, Xiyu Yi
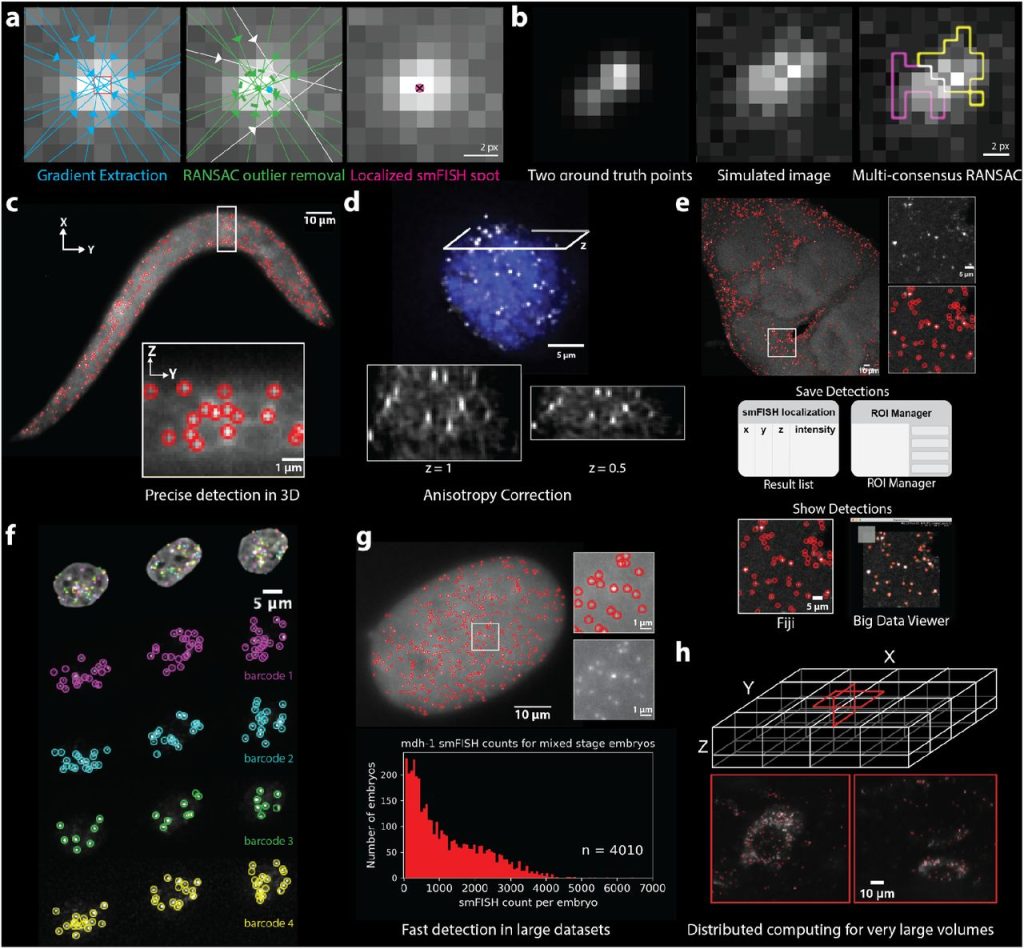
RS-FISH: Precise, interactive, fast, and scalable FISH spot detection. Ella Bahry, Laura Breimann, Marwan Zouinkhi, Leo Epstein, Klim Kolyvanov, Xi Long, Kyle I S Harrington, Timothée Lionnet, Stephan Preibisch


 (2 votes, average: 1.00 out of 1)
(2 votes, average: 1.00 out of 1)