Microscopy preprints – New tools and techniques
Posted by FocalPlane, on 19 November 2021
Here is a curated selection of preprints published recently. In this post, we focus specifically on new imaging tools only.
Open microscopy in the life sciences: Quo Vadis? Johannes Hohlbein, Benedict Diederich, Barbora Marsikova, Emmanuel G. Reynaud, Seamus Holden, Wiebke Jahr, Robert Haase, Kirti Prakash
Development of an Ontology for an Integrated Image Analysis Platform to enable Global Sharing of Microscopy Imaging Data. Satoshi Kume, Hiroshi Masuya, Yosky Kataoka, Norio Kobayashi
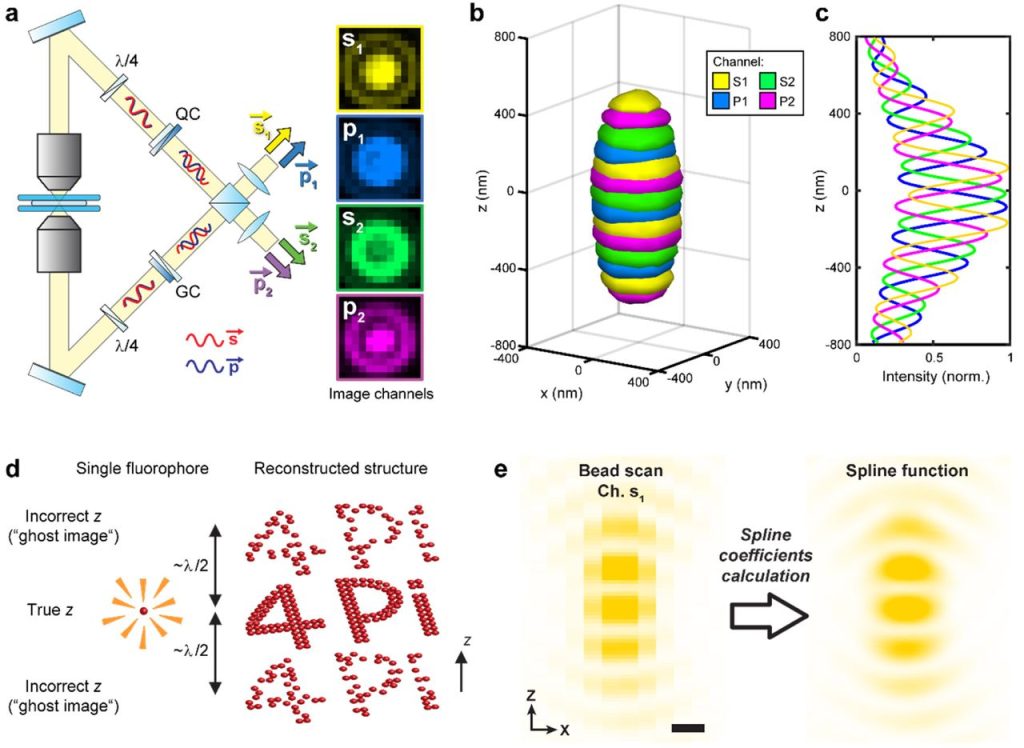
Optimal Precision and Accuracy in 4Pi-STORM using Dynamic Spline PSF Models. Mark Bates, Jan Keller-Findeisen, Adrian Przybylski, Andreas Hüper, Till Stephan, Peter Ilgen, Angel R. Cereceda Delgado, Elisa D’Este, Stefan Jakobs, Steffen J. Sahl, Stefan W. Hell
Nanoscopic resolution within a single imaging frame. Esley Torres García, Raúl Pinto Cámara, Alejandro Linares, Damián Martínez, Víctor Abonza, Eduardo Brito-Alarcón, Carlos Calcines-Cruz, Gustavo Valdés Galindo, David Torres, Martina Jabloñski, Héctor H. Torres-Martínez, José L. Martínez, Haydee O. Hernández, José P. Ocelotl-Oviedo, Yasel Garcés, Marco Barchi, Joseph G. Dubrovsky, Alberto Darszon, Mariano G. Buffone, Roberto Rodríguez Morales, Juan Manuel Rendon-Mancha, Christopher D. Wood, Armando Hernández-García, Diego Krapf, Álvaro H. Crevenna, Adán Guerrero
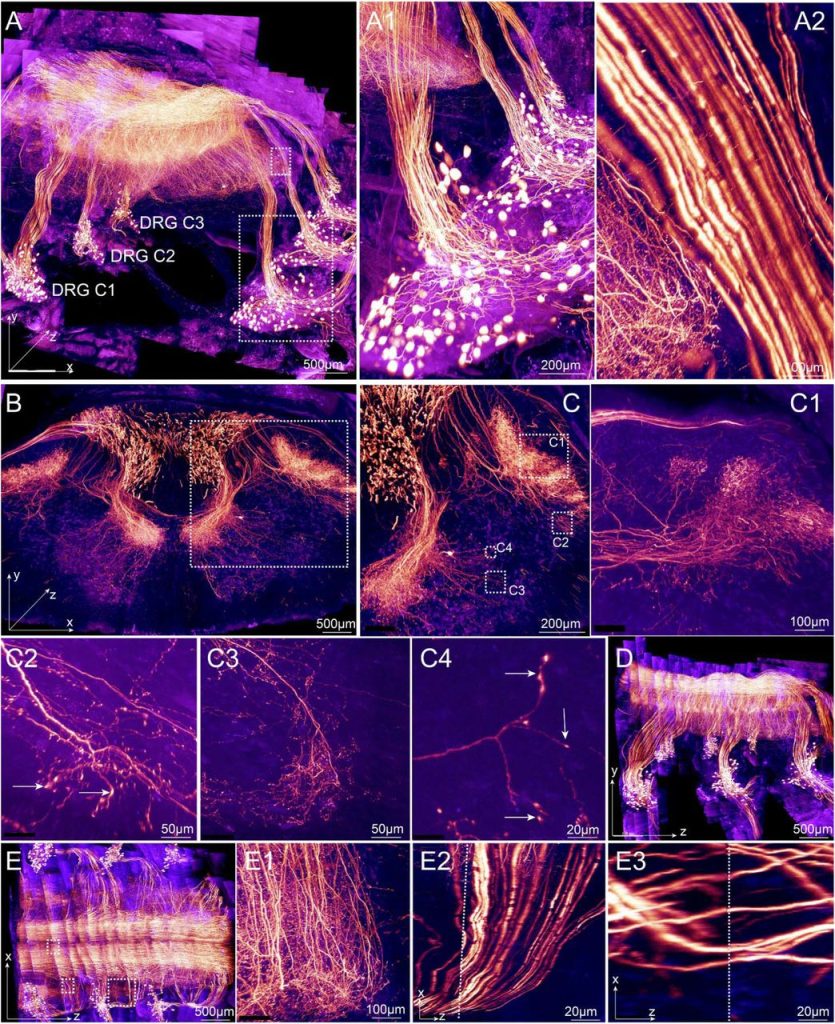
Mapping of individual sensory nerve axons from digits to spinal cord with the Transparent Embedding Solvent System. Yating Yi, Yi Men, Shiwen Zhang, Yuhong Wang, Zexi Chen, Ed Lachika, Hanchuan Peng, Woo-Ping Ge, Hu Zhao
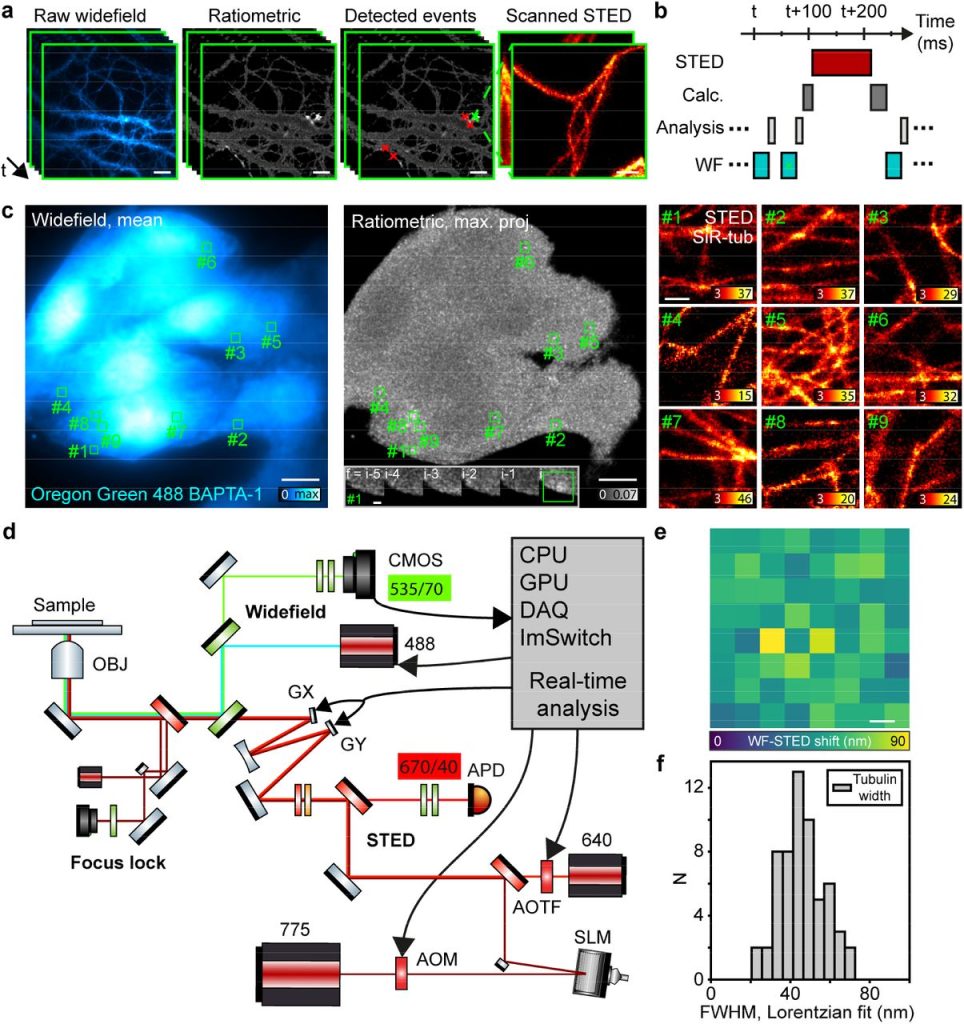
Event-triggered STED imaging. Jonatan Alvelid, Martina Damenti, Ilaria Testa
Optimal transfer functions for bandwidth-limited imaging. Sjoerd Stallinga, Antoine Delon, Jörg Enderlein
Deep-Learning Super-Resolution Microscopy Reveals Nanometer-Scale Intracellular Dynamics at the Millisecond Temporal Resolution. Rong Chen, Xiao Tang, Zeyu Shen, Yusheng Shen, Tiantian Li, Ji Wang, Binbin Cui, Yusong Guo, Shengwang Du, Shuhuai Yao
An economic, square-shaped flat-field illumination module for TIRF-based super-resolution microscopy. Jeff Y.L. Lam, Yunzhao Wu, Eleni Dimou, Ziwei Zhang, Matthew R. Cheetham, Markus Körbel, Zengjie Xia, David Klenerman, John S.H. Danial
The BrightEyes-TTM: an Open-Source Time-Tagging Module for Single-Photon Microscopy. Alessandro Rossetta, Eli Slenders, Mattia Donato, Eleonora Perego, Francesco Diotalevi, Luca Lanzanó, Sami Koho, Giorgio Tortarolo, Marco Crepaldi, Giuseppe Vicidomini
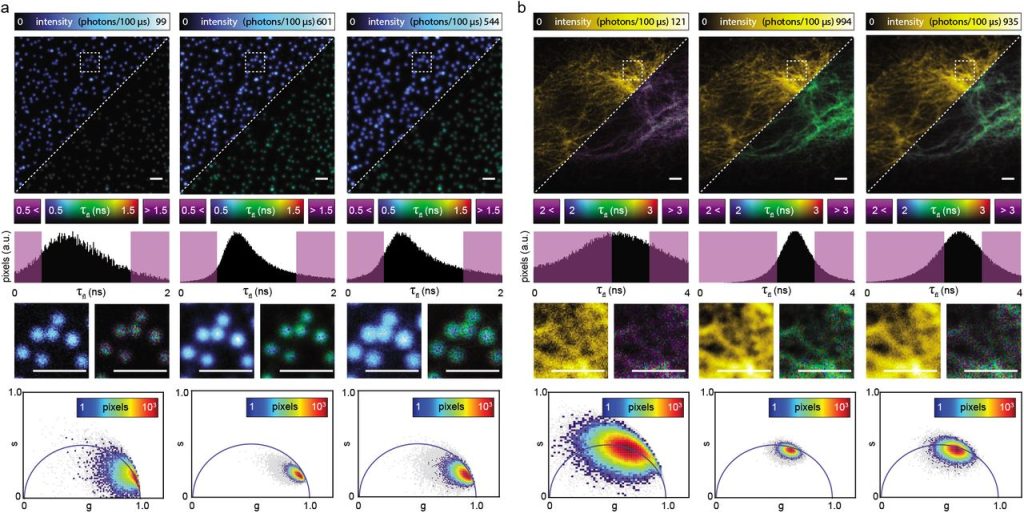
Development of ultrafast camera-based imaging of single fluorescent molecules and live-cell PALM. Takahiro K. Fujiwara, Shinji Takeuchi, Ziya Kalay, Yosuke Nagai, Taka A. Tsunoyama, Thomas Kalkbrenner, Kokoro Iwasawa, Ken P. Ritchie, Kenichi G.N. Suzuki, Akihiro Kusumi
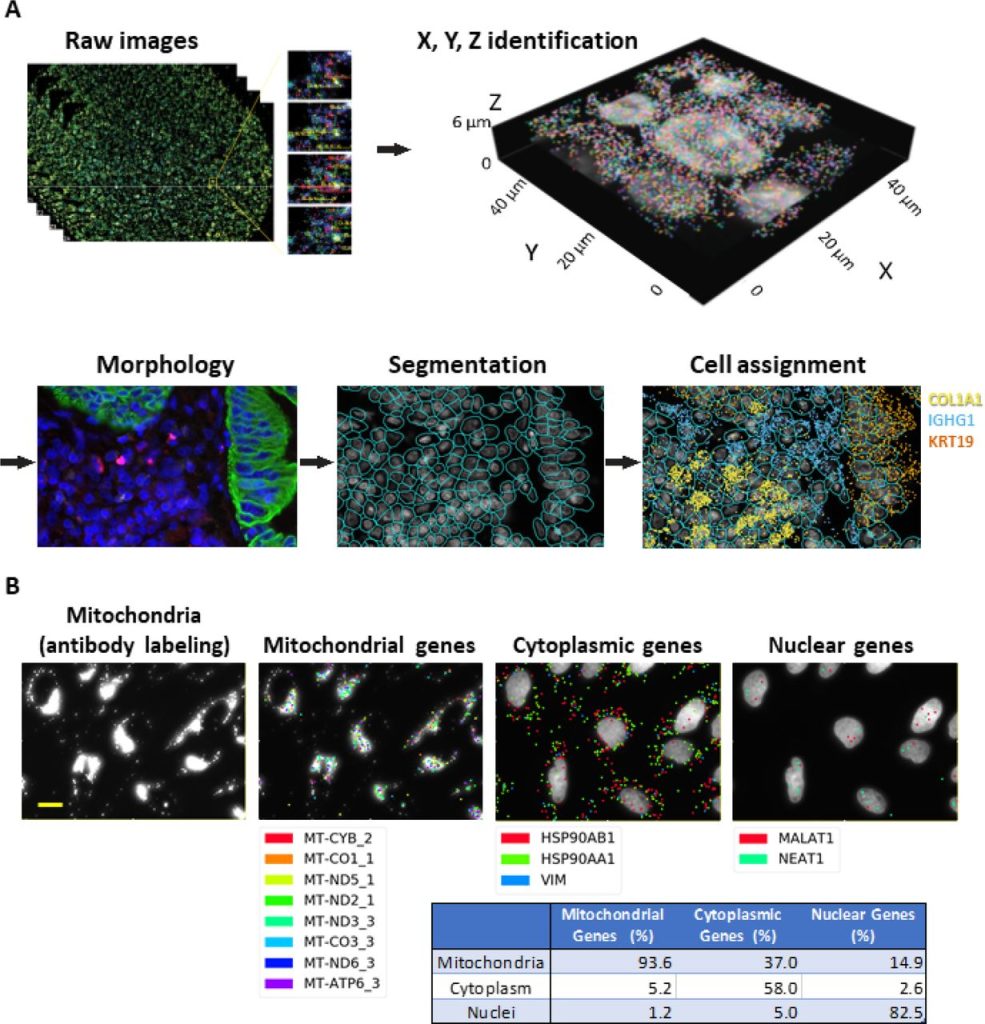
High-Plex Multiomic Analysis in FFPE Tissue at Single-Cellular and Subcellular Resolution by Spatial Molecular Imaging. Shanshan He, Ruchir Bhatt, Brian Birditt, Carl Brown, Emily Brown, Kan Chantranuvatana, Patrick Danaher, Dwayne Dunaway, Brian Filanoski, Ryan G. Garrison, Gary Geiss, Mark T. Gregory, Margaret L. Hoang, Emily E. Killingbeck, Tae Kyung Kim, Youngmi Kim, Mithra Korukonda, Alecksandr Kutchma, Erica Lee, Zachary R. Lewis, Yan Liang, Jeffrey S. Nelson, Giang Ong, Evan Perillo, Joseph Phan, Tien Phan-Everson, Erin Piazza, Tushar Rane, Zachary Reitz, Michael Rhodes, Alyssa Rosenbloom, David Ross, Hiromi Sato, Aster W. Wardhani, Corey Williams-Wietzikoski, Lidan Wu, Joseph M. Beechem
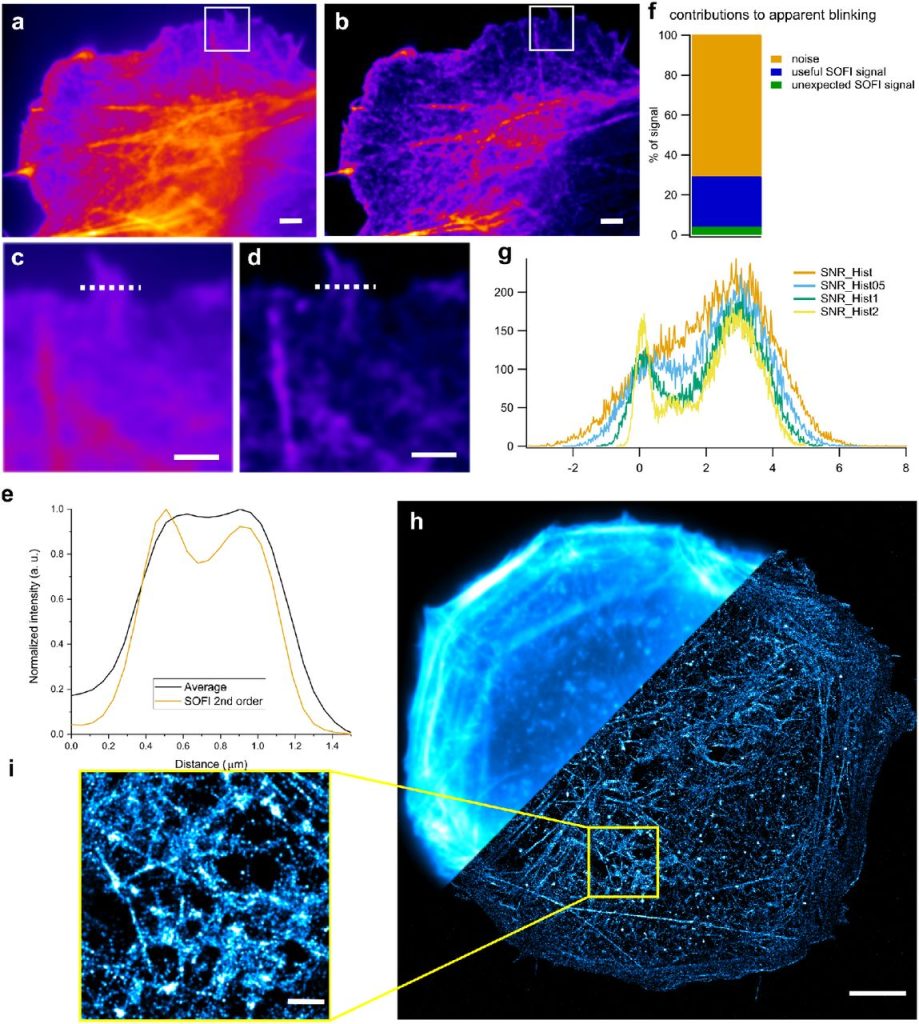
PySOFI: an open source Python package for SOFI. Yuting Miao, Shimon Weiss, Xiyu Yi
Sodium sulfite as a switching agent for single molecule based super-resolution optical microscopy. Anders Kokkvoll Engdahl, Oleg Grauberger, Mark Schüttpelz, Thomas Huser
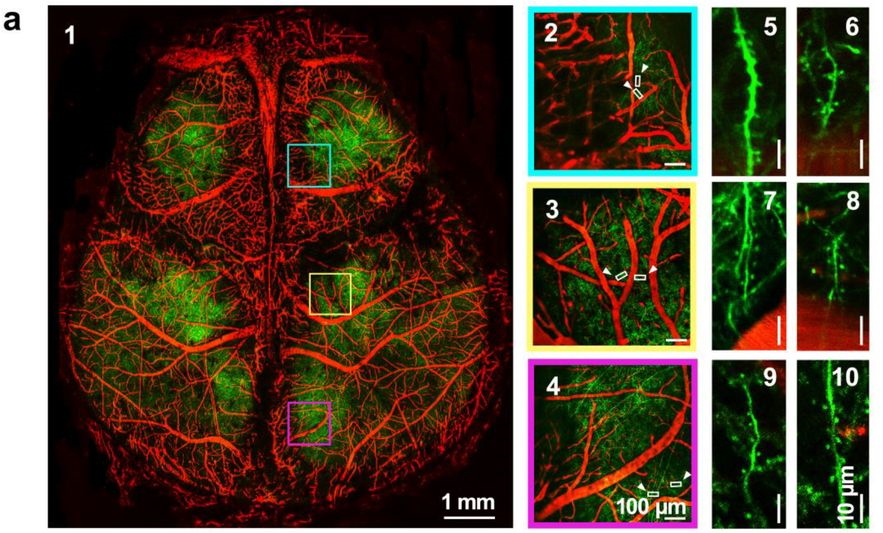
Solid optical clearing agents based through-Intact-Skull (TIS) window technique for long-term observation of cortical structure and function in mice. Dong-Yu Li, Zheng-Wu Hu, Hequn Zhang, Liang Zhu, Yin Liu, Ting-Ting Yu, Jing-Tan Zhu, Wang Xi, Jun Qian, Dan Zhu
Mod3D: A Low-Cost, Flexible Modular System of Live-Cell Microscopy Chambers and Holders. C. Barba Bazan, S. Goss, C. Peng, N. Begeja, CE. Suart, K. Neuman, Ray Truant
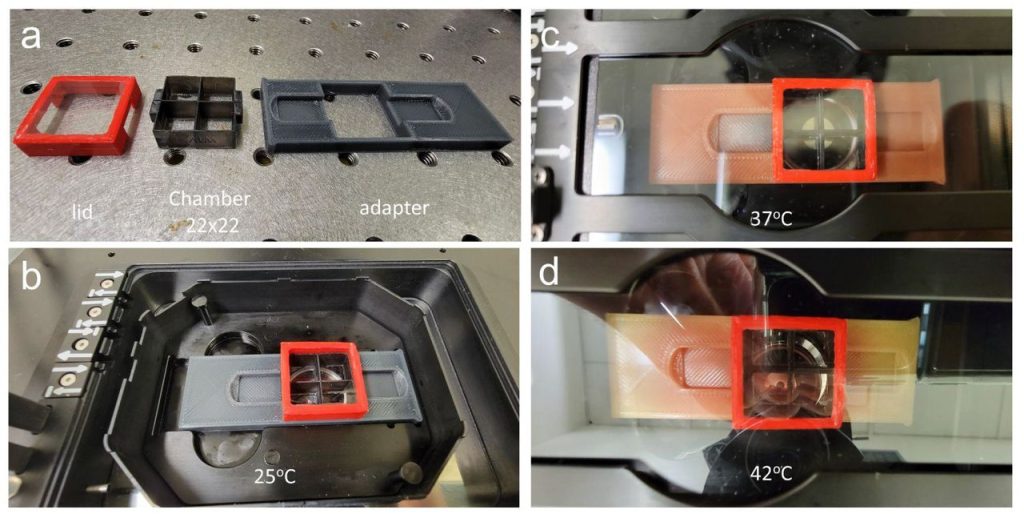
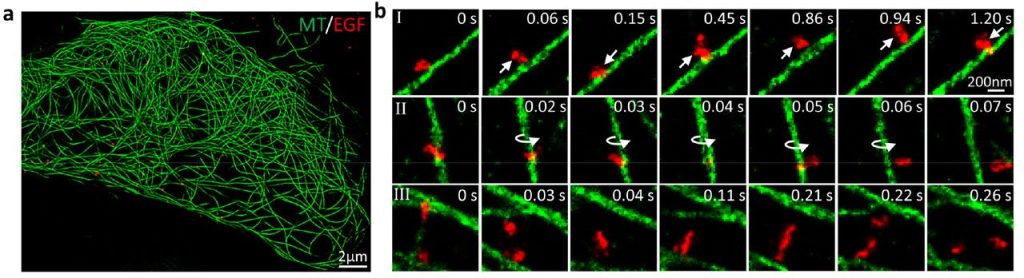
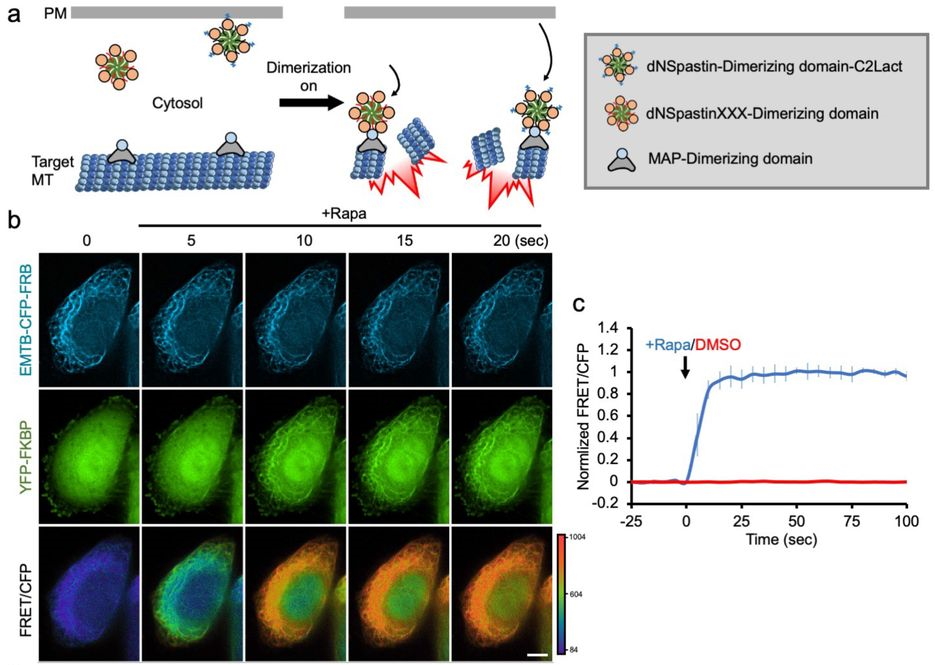
Precise control of microtubule disassembly in living cells. Grace Y. Liu, Shiau-Chi Chen, Kritika Shaiv, Shi-Rong Hong, Wen-Ting Yang, Shih-Han Huang, Ya-Chu Chang, Hsuan Cheng, Yu-Chun Lin
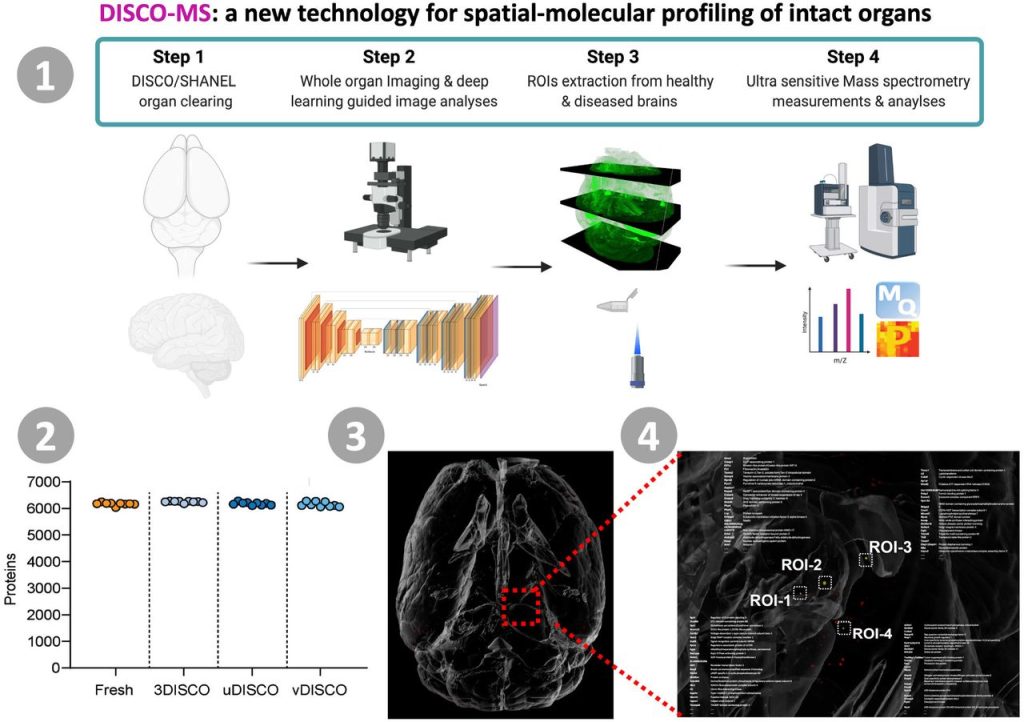
Proteomics of spatially identified tissues in whole organs. Harsharan Singh Bhatia, Andreas-David Brunner, Zhouyi Rong, Hongcheng Mai, Marvin Thielert, Rami Al-Maskari, Johannes Christian Paetzold, Florian Kofler, Mihail Ivilinov Todorov, Mayar Ali, Muge Molbay, Zeynep Ilgin Kolabas, Doris Kaltenecker, Stephan Müller, Stefan F. Lichtenthaler, Bjoern H. Menze, Fabian J. Theis, Matthias Mann, Ali Ertürk
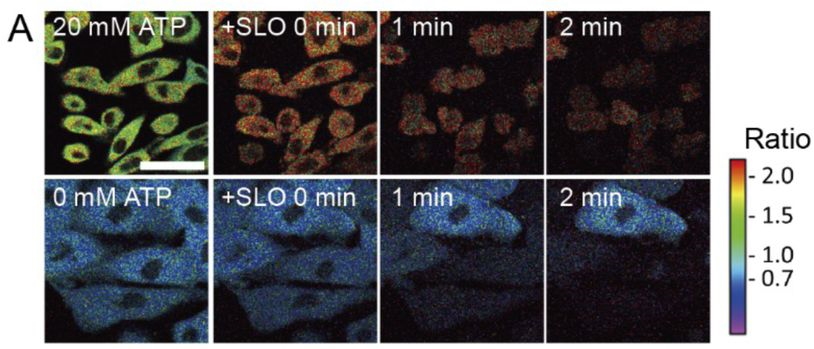
Live cell imaging of metabolic heterogeneity by quantitative fluorescent ATP indicator protein, QUEEN-37C. Hideyuki Yaginuma, Yasushi Okada
Tuning the sensitivity of genetically encoded fluorescent potassium indicators through structure-guided and genome mining strategies. Cristina C. Torres Cabán, Minghan Yang, Cuixin Lai, Lina Yang, Fedor Subach, Brian O. Smith, Kiryl D. Piatkevich, Edward S. Boyden
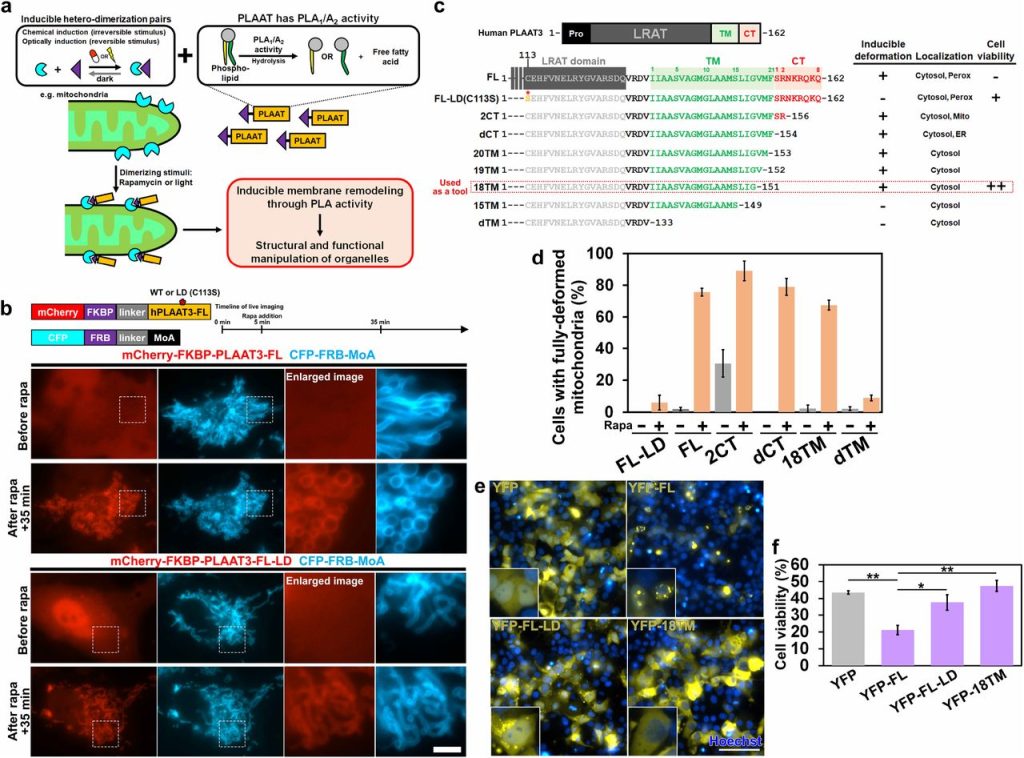
Defunctionalizing Intracellular Organelles with Genetically-Encoded Molecular Tools Based on Engineered Phospholipase A/Acyltransferases (PLAATs). Satoshi Watanabe, Yuta Nihongaki, Kie Itoh, Shigeki Watanabe, Takanari Inoue
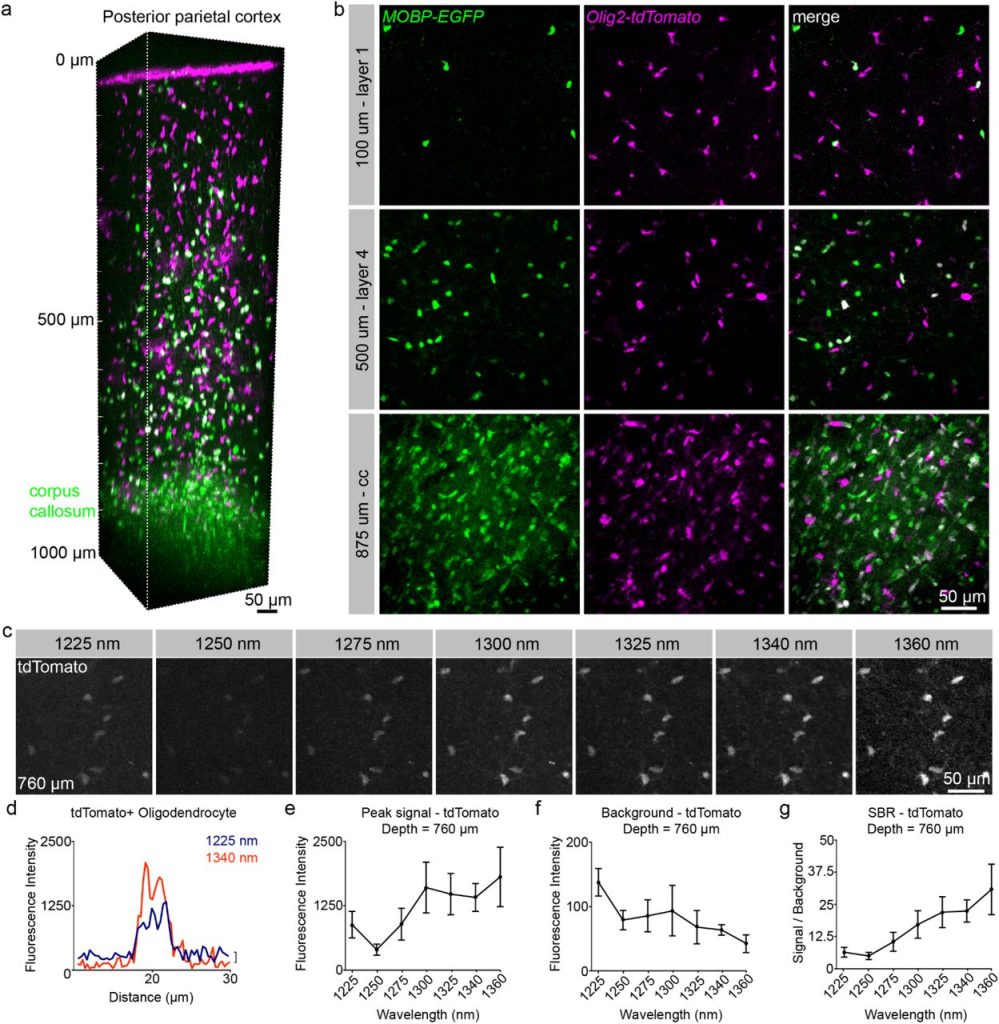
Characterization of red fluorescent reporters for dual-color in vivo three-photon microscopy. Michael A. Thornton, Gregory L. Futia, Michael E. Stockton, Baris N. Ozbay, Karl Kilborn, Diego Restrepo, Emily A. Gibson, Ethan G. Hughes
Precision localization of cellular proteins with fluorescent Fab-based probes. F. Liccardo, M. Lo Monte, B. Corrado, M. Veneruso, S. Celentano, M.R. Coscia, G. Coppola, P. Pucci, G. Palumbo, A. Luini, M. Lampe, V.M. Marzullo
IntAct: a non-disruptive internal tagging strategy to study actin isoform organization and function. M.C. van Zwam, W. Bosman, W. van Straaten, S. Weijers, E. Seta, B. Joosten, K. van den Dries
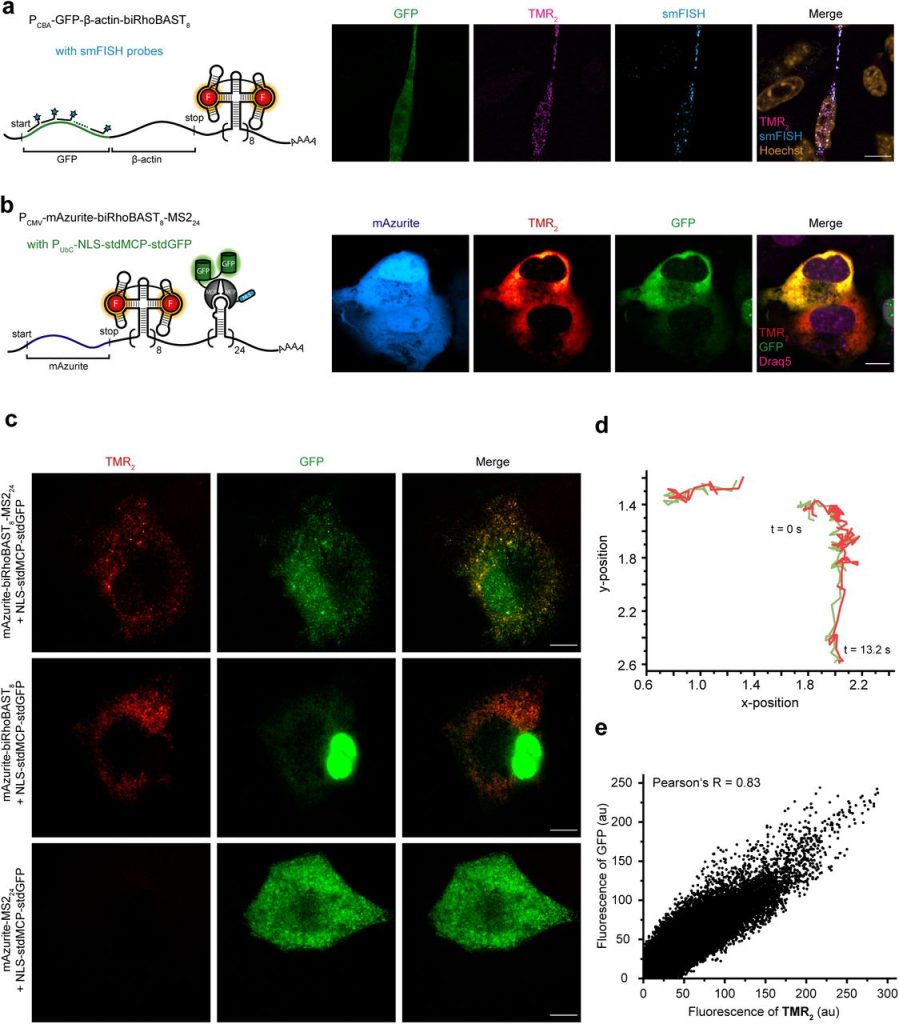
Bright, fluorogenic and photostable avidity probes for RNA imaging. Bastian Bühler, Anja Benderoth, Daniel Englert, Franziska Grün, Janin Schokolowski, Andres Jäschke, Murat Sunbul
Multifunctional Cre-dependent transgenic mice for high-precision all-optical interrogation of neural circuits. Hayley A. Bounds, Masato Sadahiro, William D. Hendricks, Marta Gajowa, Ian Antón Oldenburg, Karthika Gopakumar, Daniel Quintana, Bosiljka Tasic, Tanya L. Daigle, Hongkui Zeng, Hillel Adesnik
Sensitivity optimization of a rhodopsin-based fluorescent voltage indicator. Ahmed S Abdelfattah, Jihong Zheng, Daniel Reep, Getahun Tsegaye, Arthur Tsang, Benjamin J Arthur, Monika Rehorova, Carl VL Olson, Yi-Chieh Huang, Yichun Shuai, Minoru Koyama, Maria V Moya, Timothy D Weber, Andrew L Lemire, Christopher A Baker, Natalie Falco, Qinsi Zheng, Jonathan B Grimm, Mighten C Yip, Deepika Walpita, Craig R Forest, Martin Chase, Luke Campagnola, Gabe Murphy, Allan M Wong, Jerome Mertz, Michael N Economo, Glenn Turner, Bei-Jung Lin, Tsai-Wen Chen, Ondrej Novak, Luke D Lavis, Karel Svoboda, Wyatt Korff, Eric R Schreiter, Jeremy P Hasseman, Ilya Kolb


 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)