DeepImageJ: Deep learning in bioimage analysis for dummies
Posted by Estibaliz Gómez-de-Mariscal, on 2 December 2021
Written by Estibaliz Gómez-de-Mariscal, Cristina de-la-Torre-Gutiérrez, Arrate Muñoz-Barrutia, and Daniel Sage
Do you invest significant time looking for tools to analyze your microscopy images? Probably you have already heard about the extraordinary capabilities of deep learning (DL) to reveal complex visual structures in images. If you are a biologist practicing daily ImageJ, you may look at all these developments with an envious eye: you think that you will never have access to these technologies because the computer barrier seems impossible to overcome. What if we tell you that now you can integrate those DL models in your bioimage analysis (BIA) pipelines without previous coding knowledge? Moreover, think about a centralized repository through which you could access a collection of trained models (e.g., the BioImage Model Zoo), explore their performance, and choose the one that fits well into your analysis? This environment exists and is called deepImageJ (https://deepimagej.github.io/deepimagej/) [1].
What’s the story behind deepImageJ?
Nowadays, DL is redefining many scientific fields which are based on data, including the biomedical image analysis world. It appears with such a high level of adaptivity to the data that allows efficiently automatise the time-consuming processes (e.g., segmentation of DIC images [2] or super-resolution of specific structures [3]) or even performing tasks that otherwise would be hardly possible as the axial inpainting in non-isotropic volumes [4]. Although DL tools have shown great potential for bioimage analysis application, it has not always been as easy to deploy them in biology labs due to the computer skills requirements and the specific hardware requirements. Specifically, deploying DL models needs coding expertise, machine learning proficiency, and image processing background.
Arrate Muñoz-Barrutia (Universidad Carlos III de Madrid, Spain) and Daniel Sage (École Polytechnique Fédérale de Lausanne,Switzerland) were aware of the huge gap between the DL Python libraries (TensorFlow and PyTorch) and the most commonly used tool in bioimage analysis: ImageJ [5] (https://imagej.net/). Thus, they conceived the idea of running trained DL models directly in ImageJ in one click, as a standard plugin of ImageJ. So, deepImageJ was born as an idea to make trained DL models, user-friendly and accessible for every scientist.
The development of deepImageJ was paved thanks to pioneer works such as CSBDeep [4] or the TensorFlow version manager [6], which were the very first tools bringing DL-related solutions to the ImageJ ecosystem. Thanks to these previous works and focusing on becoming as general as possible, deepImageJ managed to release an ImageJ plugin agnostic to image processing tasks, offering a unified and generic way to access pre-trained models. In other words, deepImageJ transformed the deployment of DL image processing workflows into a regular ImageJ plugin that life-scientists could include in their daily routine. Because deepImageJ aimed to make DL usable by non-experts, a massive interaction with life-scientists, bioimage-analysts, computer scientists and engineers was one of the best sources to collect feedback, and achieve a competitive, mature and versatile tool.
How does deepImageJ support bioimage analysis?
Several DL-based image processing methods are published every year, but barely a few are directly reusable in laboratory practices. DeepImageJ appears to keep alive these novel methods by linking DL and life-scientists. Moreover, deepImageJ’s operability is independent of the model’s architecture. So, as you might have figured out, it allows for image-to-image (i.e., denoising) and image-to-void (i.e., image classification) processing.
DeepImageJ lives in the ImageJ ecosystem by providing a user-friendly plugin that can be installed through the update sites (https://imagej.net/update-sites/). DeepImageJ is macro recordable and macro callable. That is, you can use it to easily design bioimage analysis pipelines in which images are processed with a selected DL model.
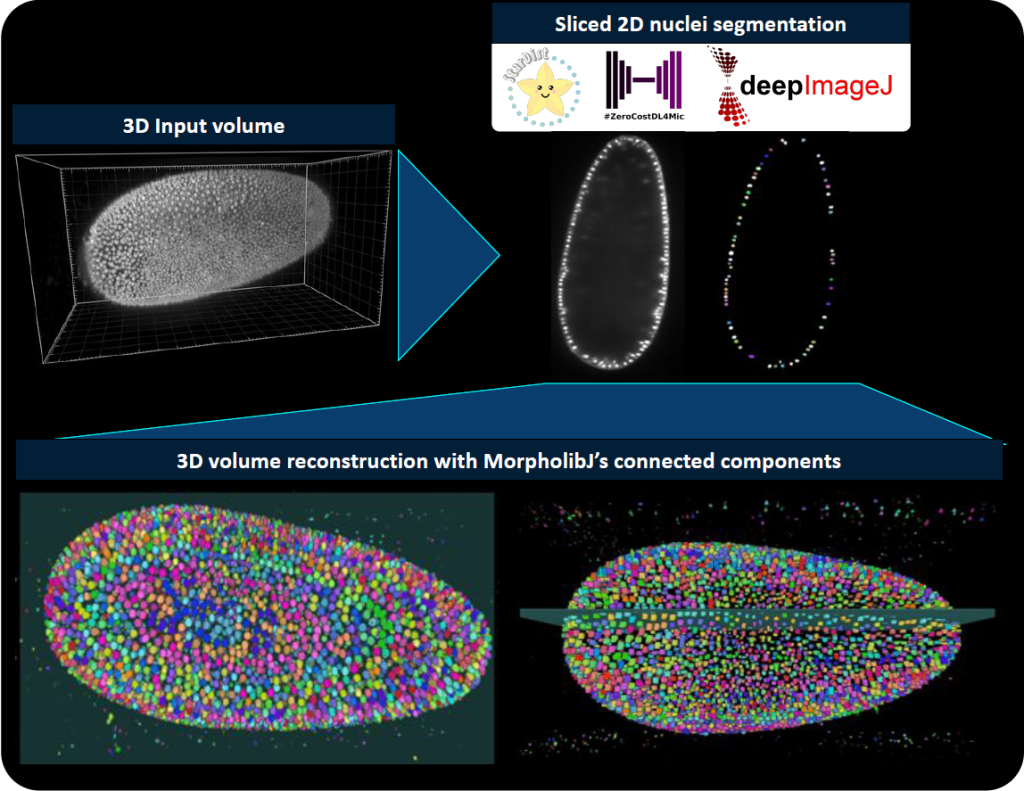
How do we bridge deep learning and life-scientists?
Practically speaking, DL models are ensembles of mathematical operations (i.e., neural network architecture) that process an image and provide an output as a result, which can be for example, an image, a numerical value, or a list of features. If the DL model is properly loaded on a system, knowing the mathematical properties of the input image and the output, and how they should be handled (pre- and post-processing steps) suffices to define the rules to deploy the DL image processing. All this information is usually controlled by the model developer. DeepImageJ defined for the first time, a model format that gathers all this information, usually controlled by the model developer, and it is still adapted to different image processing tasks or DL models. Thus, deepImageJ integrates in a unified manner the pre-processing, prediction and post-processing, everything in a click. Such model format, now updated to the BioImage Model Zoo (https://bioimage.io) format, is what makes deepImageJ a generic plugin for DL and therefore, it allows life-scientists to access a long list of different DL models.
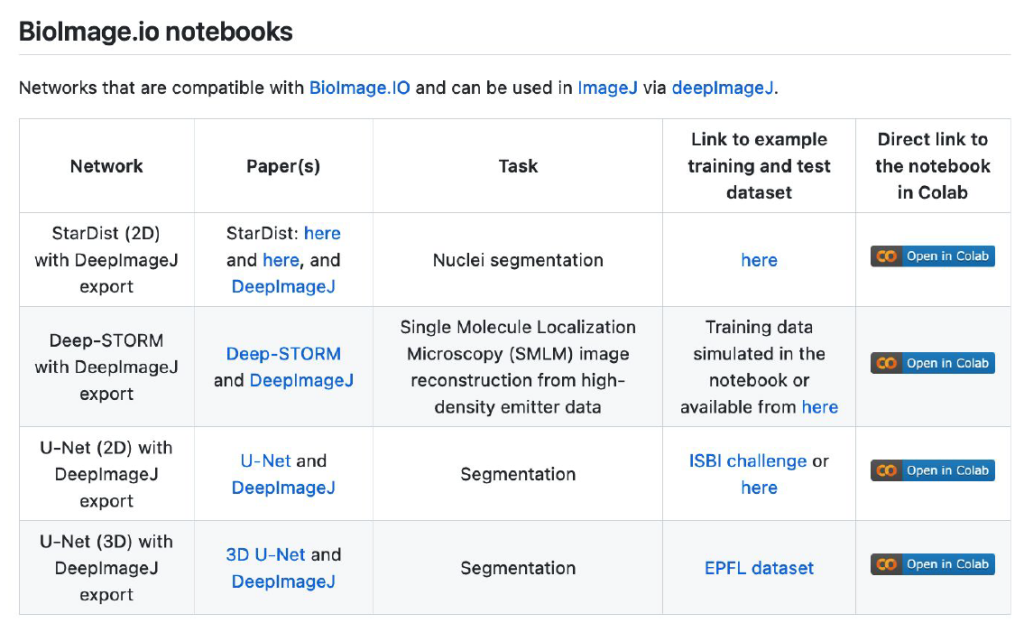
Yet another drawback of DL models is the need to be (re-)trained. Most DL models fail to accurately process new images that have a data shift compared to the trained images l (e.g., change of scale, brightfield microscopy and fluorescence microscopy images). To allow model training and fine-tuning of a model on certain specific data, some of the models in ZeroCostDL4Mic [8] can be exported into the deepImageJ format. Because ZeroCostDL4Mic lets non-expert researchers train state-of-the-art models as the U-Net [2], such synergy makes it possible to get suitable models for specific bioimage analysis tasks in ImageJ, and therefore it equips researchers with yet a more versatile tool.
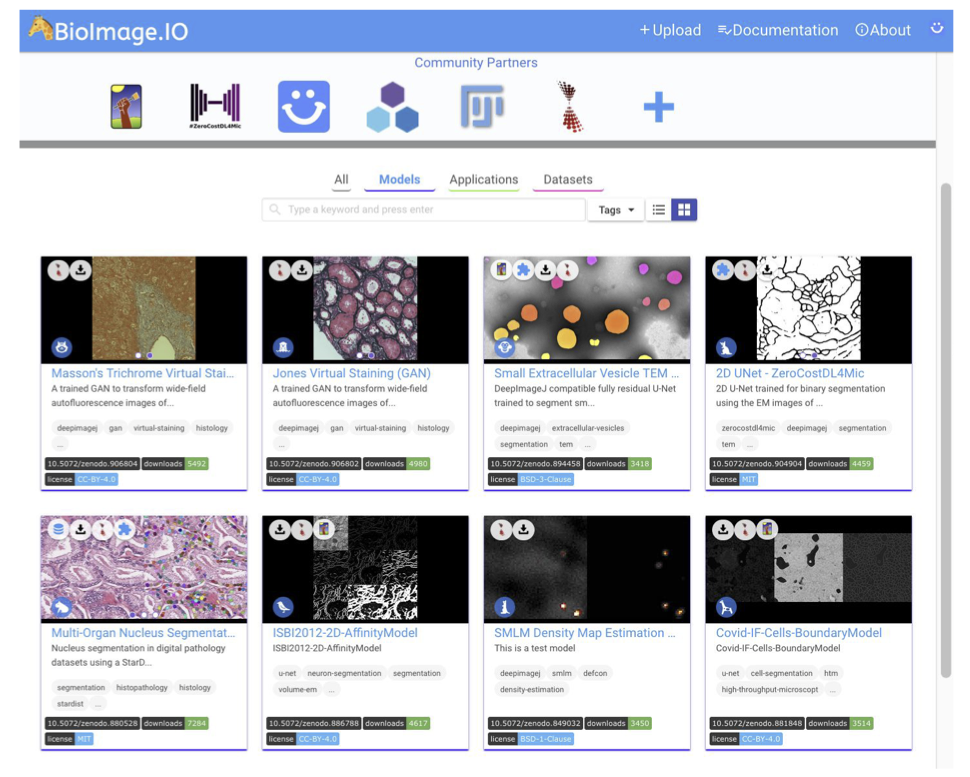
Of course, one should also ask about a potential source to access pre-trained DL models and how to disseminate all these powerful tools. The first way envisioned by deepImageJ collaborators is the spread of models from image processing challenges such as it has been done with the MU-Lux-CZ submission in the Cell Tracking Challenge [11]. But even more exciting, there is the new-born BioImage Model Zoo (https://bioimage.io/) community-driven AI model repository, which encompasses a large collection of trained DL models for bioimage analysis that can be used in Ilastik [12], DeepImageJ, ImJoy [13], ZeroCostDL4Mic [8] or CSBDeep [4]. In this case, the deepImageJ model format has been updated to follow the specifications defined by all the community partners that form this initiative.
So, how can we use deepImageJ?
There was once a scientist that was tired of segmenting cells manually to carry out their research. She/He had heard about DL and how other laboratories are benefiting from it already to do this kind of task automatically. Finally, she/he decided to enter the BioImage Model Zoo repository to find a DL model that adjusts better to its needs. Successfully, she/he finds one DL model available that fits well to the type of cells that she/he has, so she/he decides to try it on his own by downloading it and using it in the deepImageJ plugin.
In only a few steps, this life-scientist was able to explore the Bioimage Model Zoo repository, download the DL model that she/he wants to use in deepImageJ, install it, and use it successfully in ImageJ. So, beyond its direct use, deepImageJ contributes to the spread and validation of DL models in life-science applications in order to make them visible and alive.
The big feedback and proof of DL-user-friendly tools
DeepImageJ is contributing to making DL more accessible and seeks to efficiently support life-scientists in their daily routines. Although it is a young project, it has already been published in Nature Methods, there is an upcoming book chapter about it, and has participated in more than 10 seminars and courses and different renowned conferences all around the world.
As proof of deepImageJ usability, the number of downloads has been continuously increasing during the last months, until obtaining a total of 4’000 (source: https://github.com/deepimagej/deepimagej). In other words, we discovered an increasing number of researchers keen on learning DL-based bioimage analysis and who need it in their daily practice. Do you feel you could be one of those researchers? So, do not take our word for it, experience it for yourself.

Authors
Estibaliz Gómez-de-Mariscal, PhD – Postdoctoral scholar (Universidad Carlos III de Madrid, Instituto Gulbenkian de Ciência, Henriques Laboratory)
Cristina de-la-Torre-Gutiérrez – PhD Candidate (Universidad Carlos III de Madrid, BiiG)
Arrate Muñoz-Barrutia, PhD – Full professor (Universidad Carlos III de Madrid, BiiG)
Daniel Sage, PhD — Senior Scientist (EPFL, EPFL Center for Imaging)
References
1. Gómez-de-Mariscal, E., García-López-de-Haro, C., Ouyang, W., et al., DeepImageJ: A user-friendly environment to run deep learning models in ImageJ. Nature Methods 18, 1192–1195 (2021). https://doi.org/10.1038/s41592-021-01262-9
2. Falk, T., Mai, D., Bensch, R., et al. ,U-Net: deep learning for cell counting, detection, and morphometry. Nature Methods 16, 67–70 (2019). https://doi.org/10.1038/s41592-018-0261-2
3. Nehme, E., Weiss, L.E., Michaeli, T., Schechtman, Y., Deep-STORM: super-resolution single-molecule microscopy by deep learning, Optica 5, 458-464 (2018). https://doi.org/10.1364/OPTICA.5.000458
4. Weigert, M., Schmidt, U., Boothe, T., et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nature Methods 15, 1090–1097 (2018). https://doi.org/10.1038/s41592-018-0216-7
5. Schneider, C.A., Rasband, W. & Eliceiri, K. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675 (2012). https://doi.org/10.1038/nmeth.2089
6. Schroeder, A.B., Dobson, E.T.A., Rueden, C.T., et al., The ImageJ ecosystem: Open-source software for image visualization, processing, and analysis, Protein Science, 30(1), 234-249 (2021). https://onlinelibrary.wiley.com/doi/epdf/10.1002/pro.3993
7. Tsai, H.F., Gajda, J., Sloan, T.F.W., et al., Usiigaci: Instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning, SoftwareX 9, 230-237 (2019). https://doi.org/10.1016/j.softx.2019.02.007
8. von Chamier, L., Laine, R.F., Jukkala, J., et al., Democratising deep learning for microscopy with ZeroCostDL4Mic. Nature Communications 12, 2276 (2021). https://doi.org/10.1038/s41467-021-22518-0
9. Schmidt, U., Weigert, M., Broaddus, C., Myers, G., Cell detection with star-convex polygons. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science, vol 11071. Springer, Cham, (2018). https://doi.org/10.1007/978-3-030-00934-2_30
10. Legland, D., Arganda-Carreras, I., Andrey P., MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ, Bioinformatics 32 (2016). https://doi.org/10.1093/bioinformatics/btw413
11. Ulman, V., Maška, M., Magnusson, K., et al., An objective comparison of cell-tracking algorithms. Nature Methods 14, 1141–1152 (2017). https://doi.org/10.1038/nmeth.4473
12. Berg, S., Kutra, D., Kroeger, T., et al.., Ilastik: Interactive learning and segmentation toolkit. Nature Methods 16, 1226–1232 (2019). https://doi.org/10.1038/s41592-019-0582-9
13. Ouyang, W., Mueller, F., Hjelmare, M. et al., ImJoy: an open-source computational platform for the deep learning era. Nat Methods 16, 1199–1200 (2019). https://doi.org/10.1038/s41592-019-0627-0


 (No Ratings Yet)
(No Ratings Yet)