Microscopy preprints – Bioimage analysis tools
Posted by FocalPlane, on 17 December 2021
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools only.
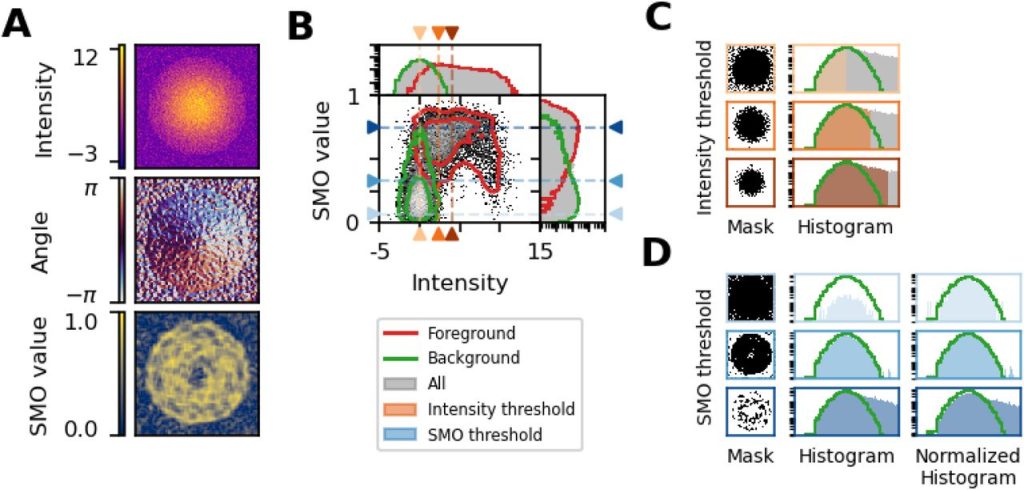
Robust and unbiased estimation of the background distribution for automated quantitative imaging. Mauro Silberberg, Hernán E. Grecco
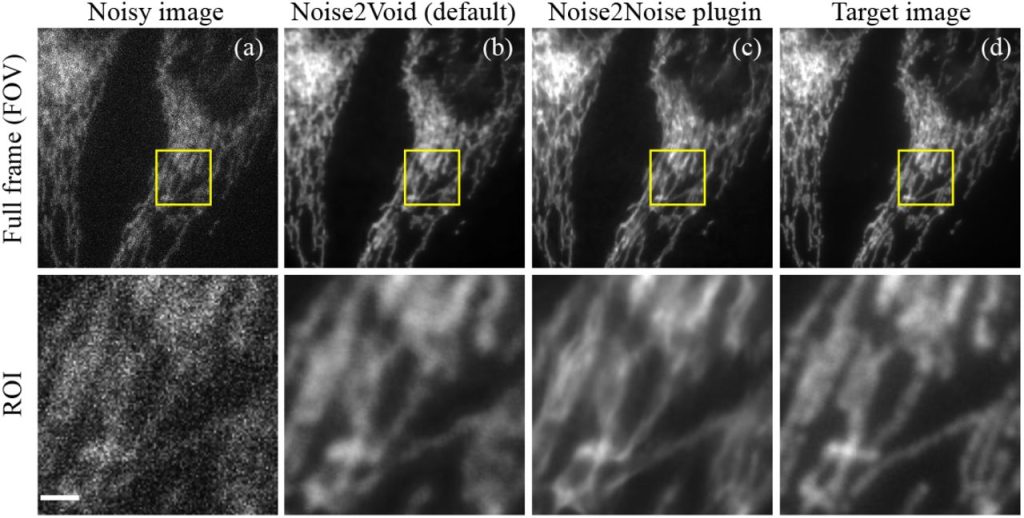
Real-time Image Denoising of Mixed Poisson-Gaussian Noise in Fluorescence Microscopy Images using ImageJ. Varun Mannam, Yide Zhang, Yinhao Zhu, Evan Nichols, Qingfei Wang, Vignesh Sundaresan, Siyuan Zhang, Cody Smith, Paul W Bohn, Scott Howard
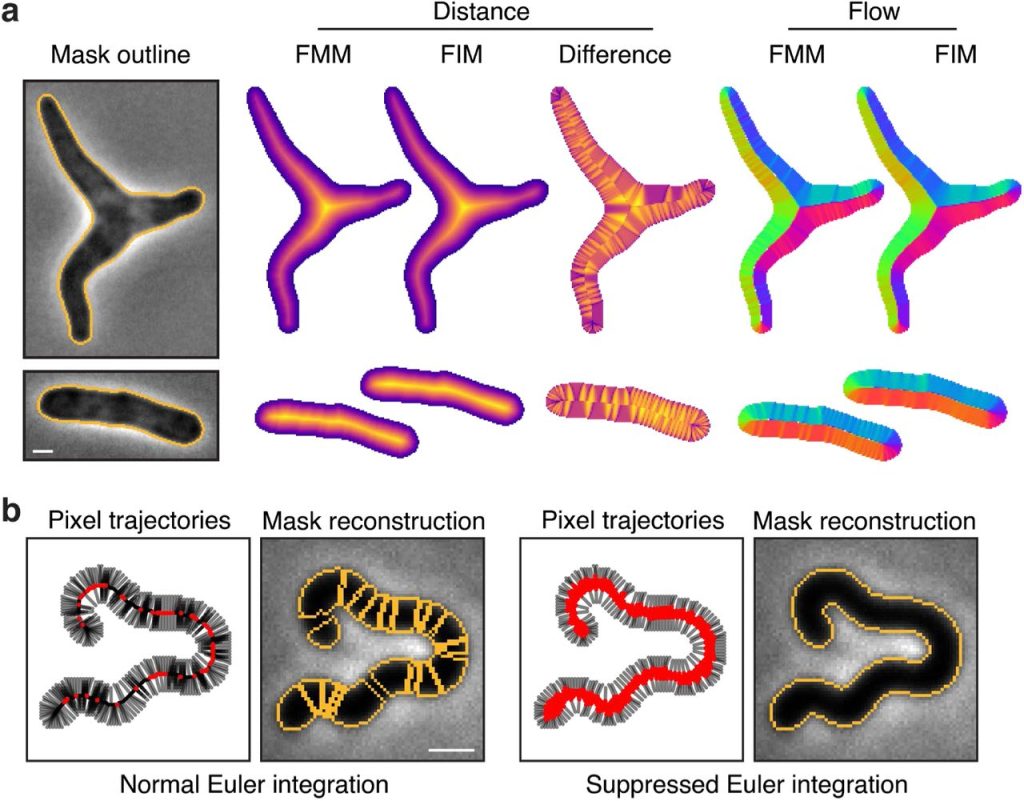
Omnipose: a high-precision morphology-independent solution for bacterial cell segmentation. Kevin J. Cutler, Carsen Stringer, Paul A. Wiggins, Joseph D. Mougous
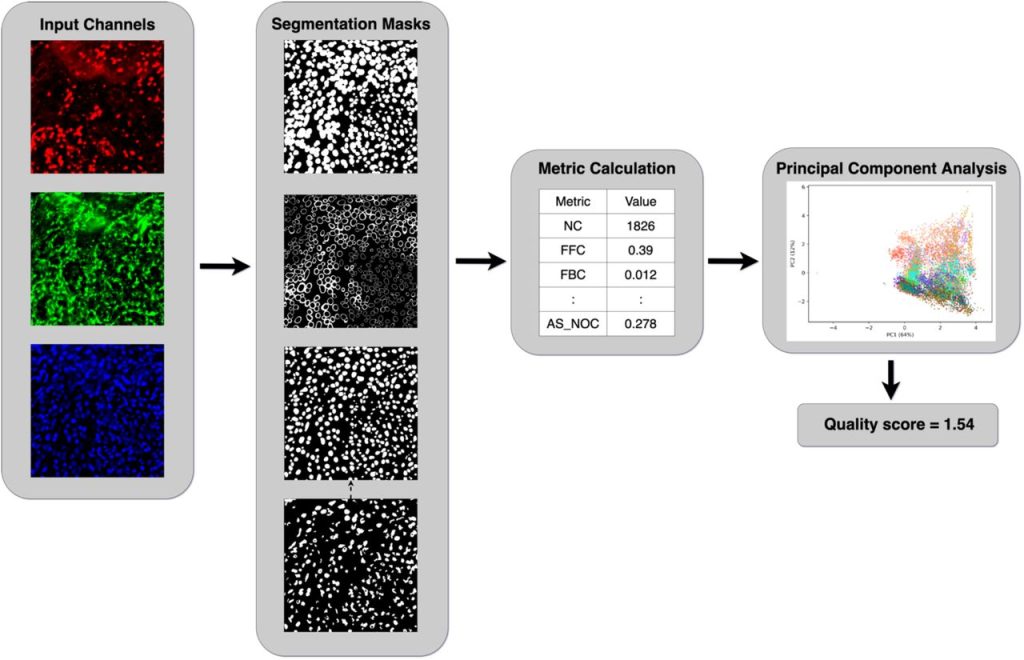
Evaluation of cell segmentation methods without reference segmentations. Haoran Chen, Robert F. Murphy
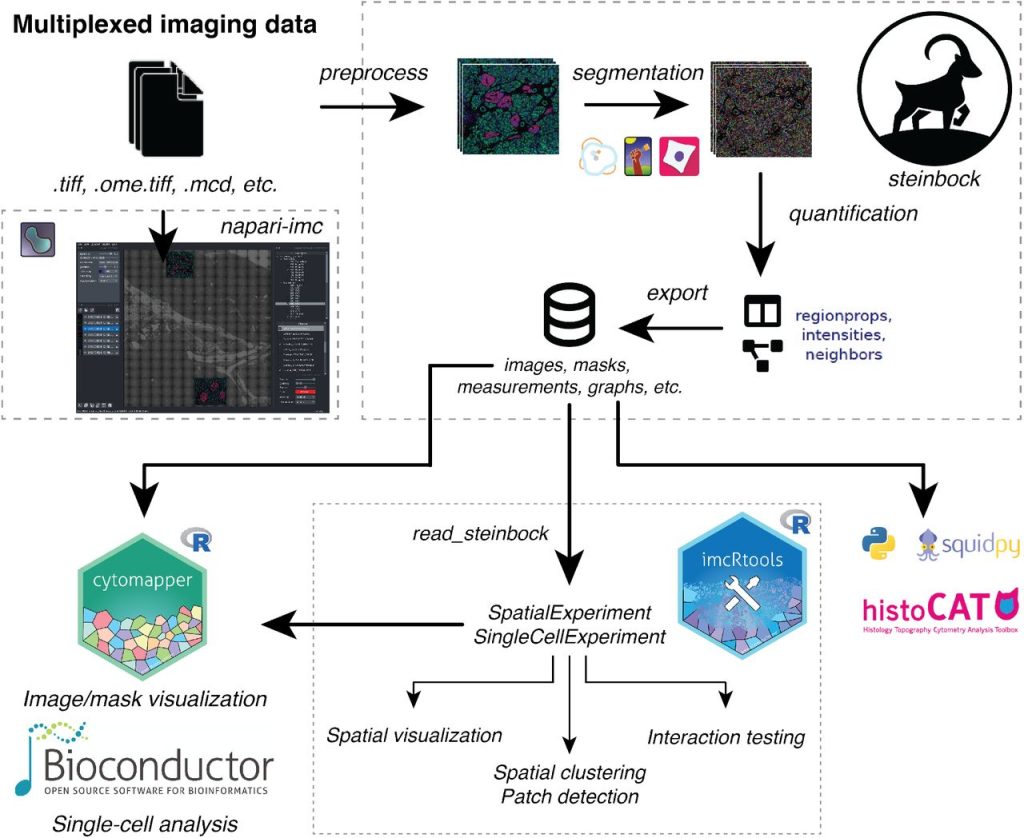
An end-to-end workflow for multiplexed image processing and analysis. Jonas Windhager, Bernd Bodenmiller, Nils Eling
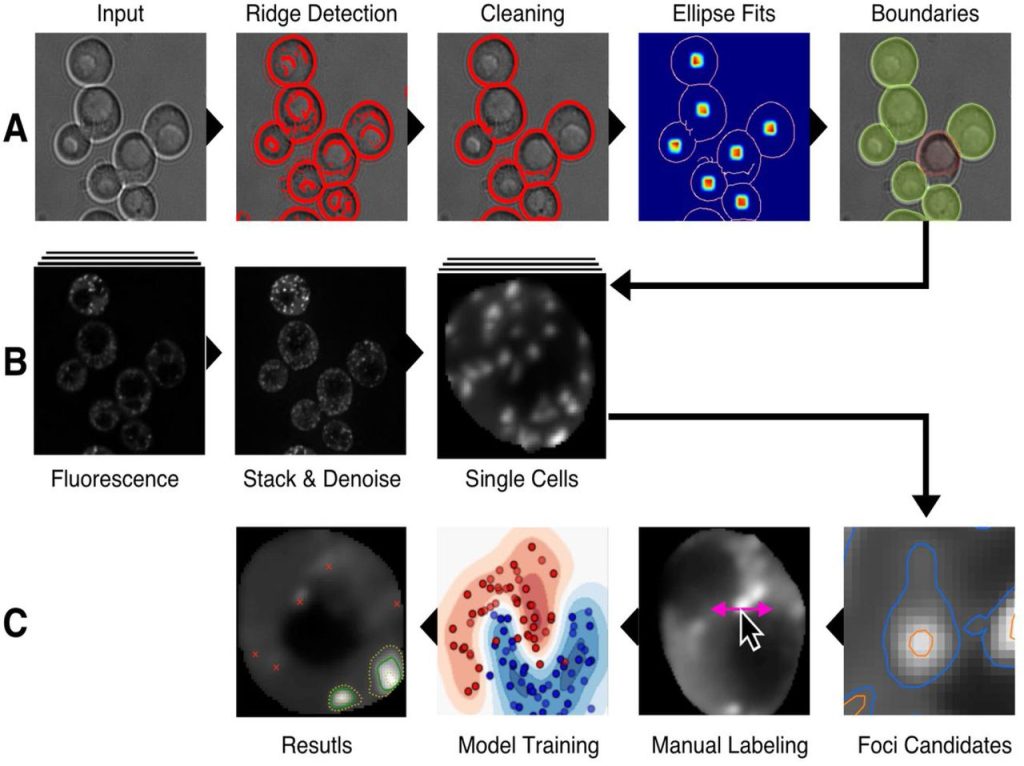
HARLEY: A Semi-Automated detection of foci in fluorescence images of yeast. Ilya Shabanov, J. Ross Buchan
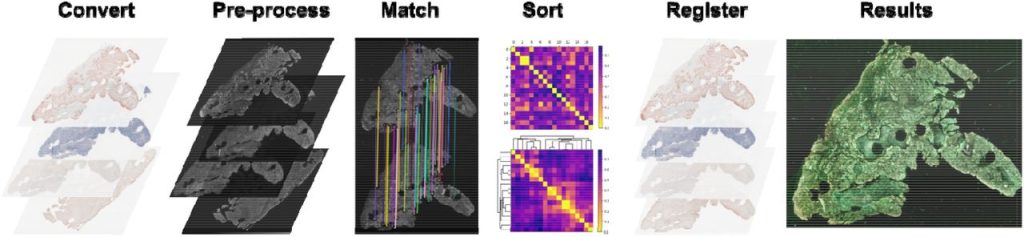
VALIS: Virtual Alignment of pathoLogy Image Series. Chandler D. Gatenbee, Ann-Marie Baker, Sandhya Prabhakaran, Robbert J. C. Slebos, Gunjan Mandal, Eoghan Mulholland, Simon Leedham, Jose R. Conejo-Garcia, Christine H. Chung, Mark Robertson-Tessi, Trevor A. Graham, Alexander R.A. Anderson
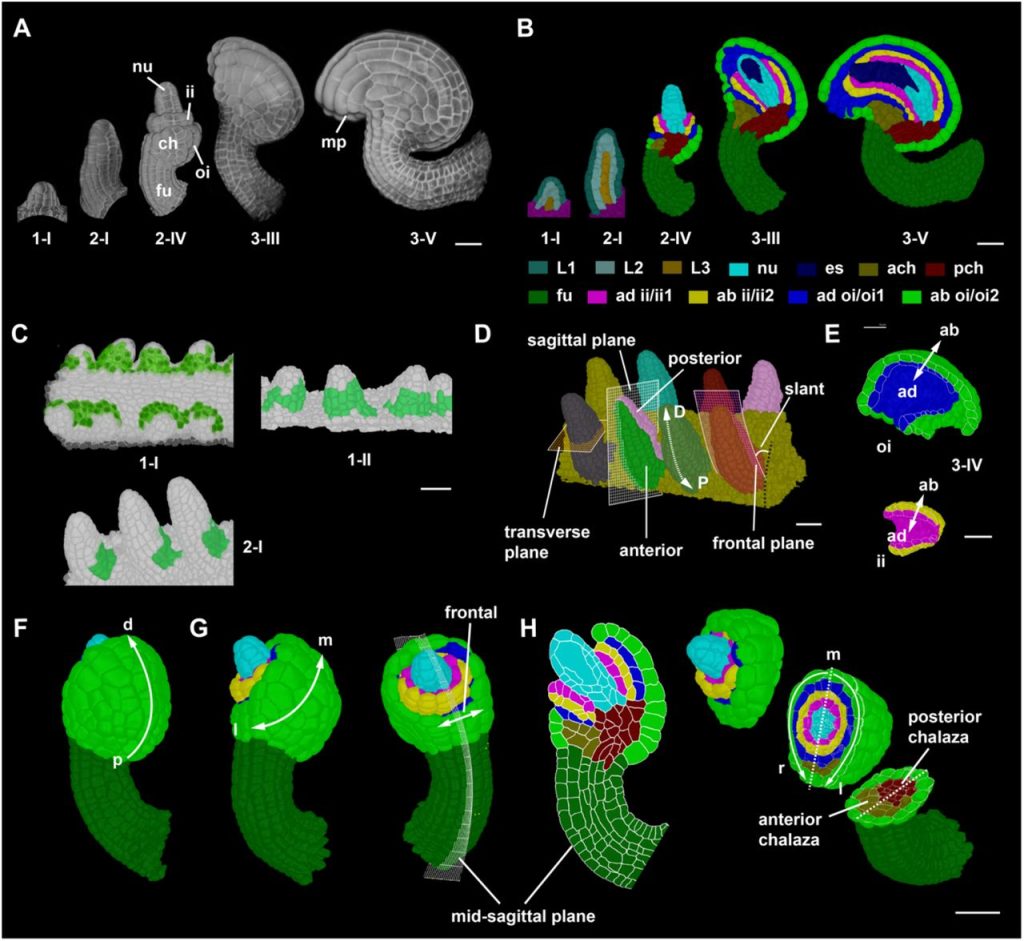
The annotation and analysis of complex 3D plant organs using 3DCoordX. Athul Vijayan, Soeren Strauss, Rachele Tofanelli, Tejasvinee Atul Mody, Karen Lee, Miltos Tsiantis, Richard S Smith, Kay Schneitz
LiveCellMiner: A New Tool to Analyze Mitotic Progression. Daniel Moreno-Andrés, Anuk Bhattacharyya, Anja Scheufen, Johannes Stegmaier
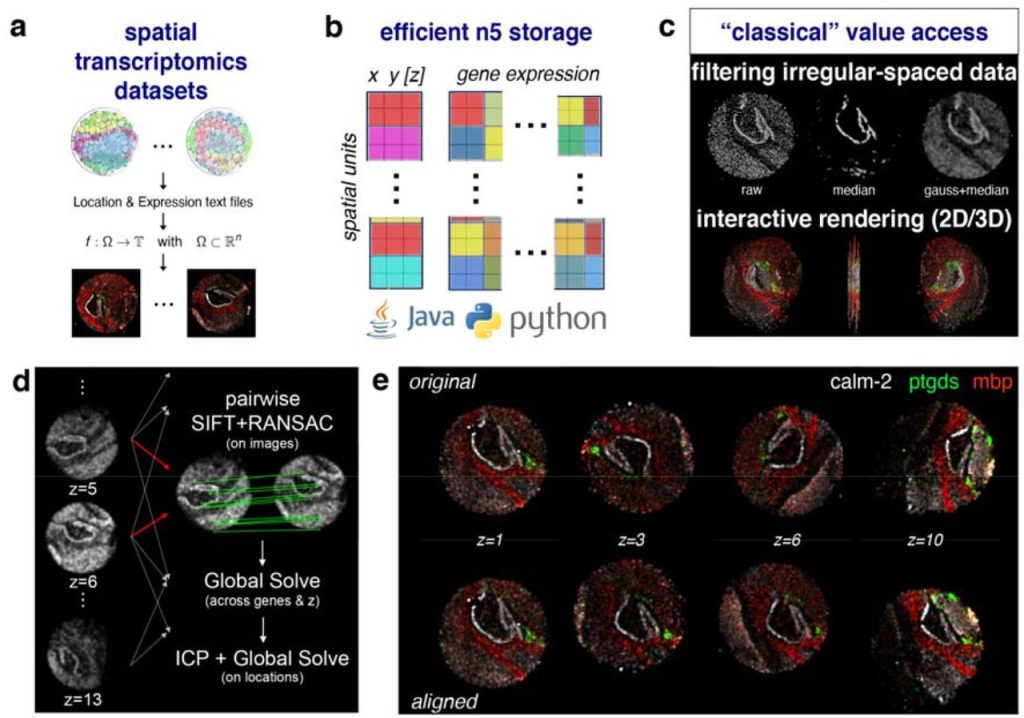
Spacemake: processing and analysis of large-scale spatial transcriptomics data. Tamas Ryszard Sztanka-Toth, Marvin Jens, Nikos Karaiskos, Nikolaus Rajewsky
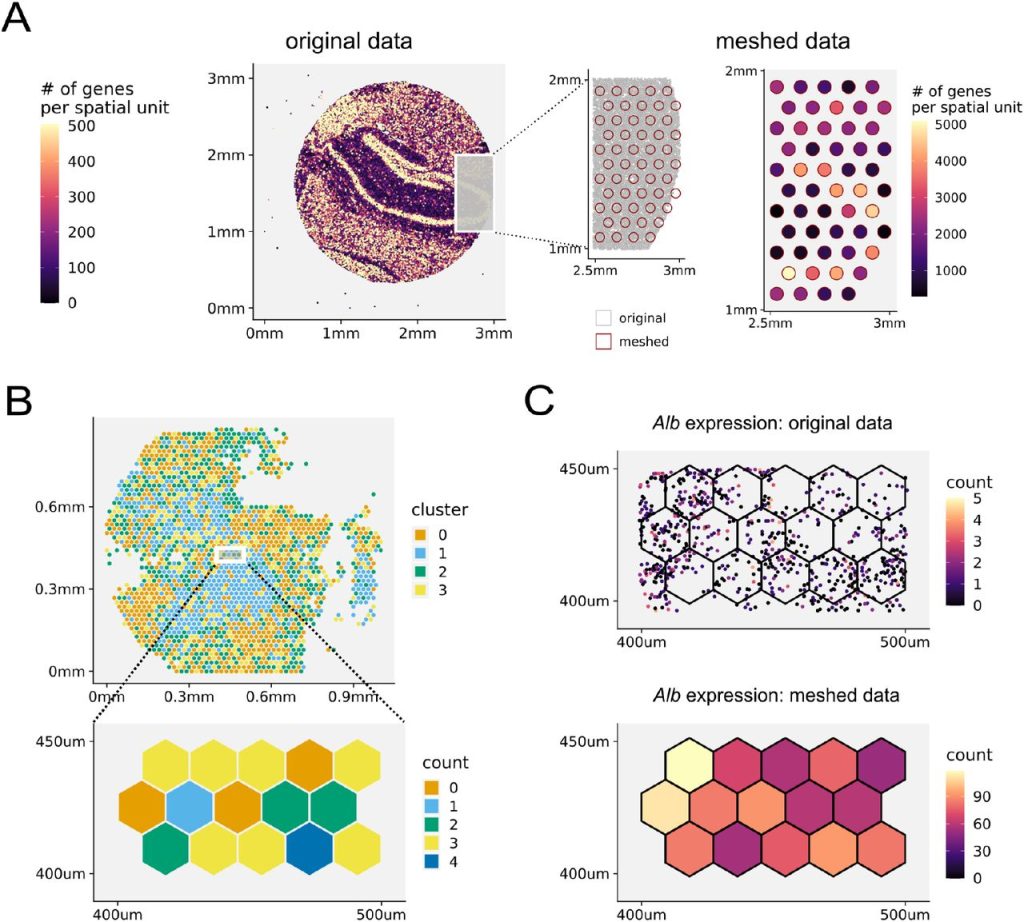
Image-based representation of massive spatial transcriptomics datasets. Stephan Preibisch, Nikos Karaiskos, Nikolaus Rajewsky
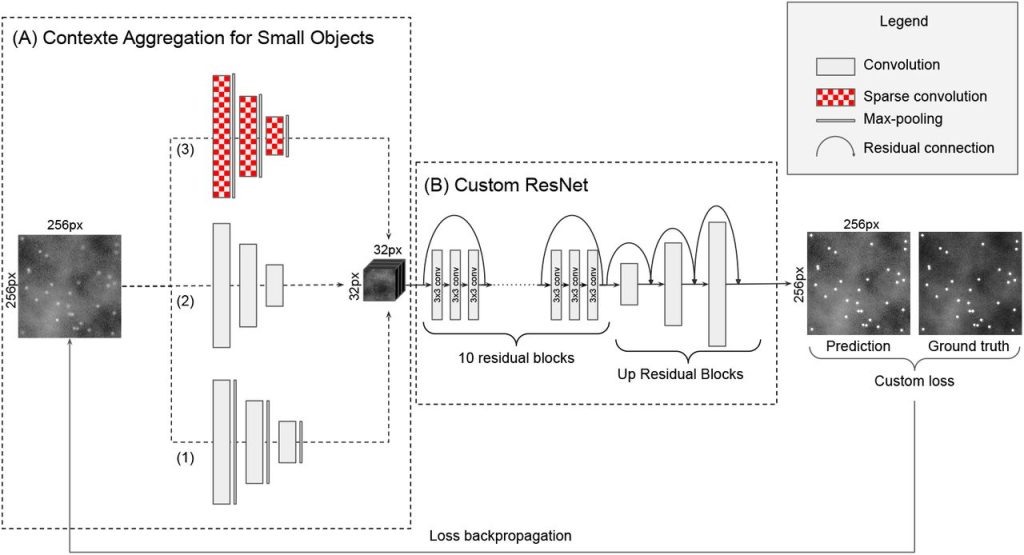
DeepSpot: a deep neural network for RNA spot enhancement in smFISH microscopy images. Emmanuel Bouilhol, Edgar Lefevre, Benjamin Dartigues, Robyn Brackin, Anca Flavia Savulescu, Macha Nikolski
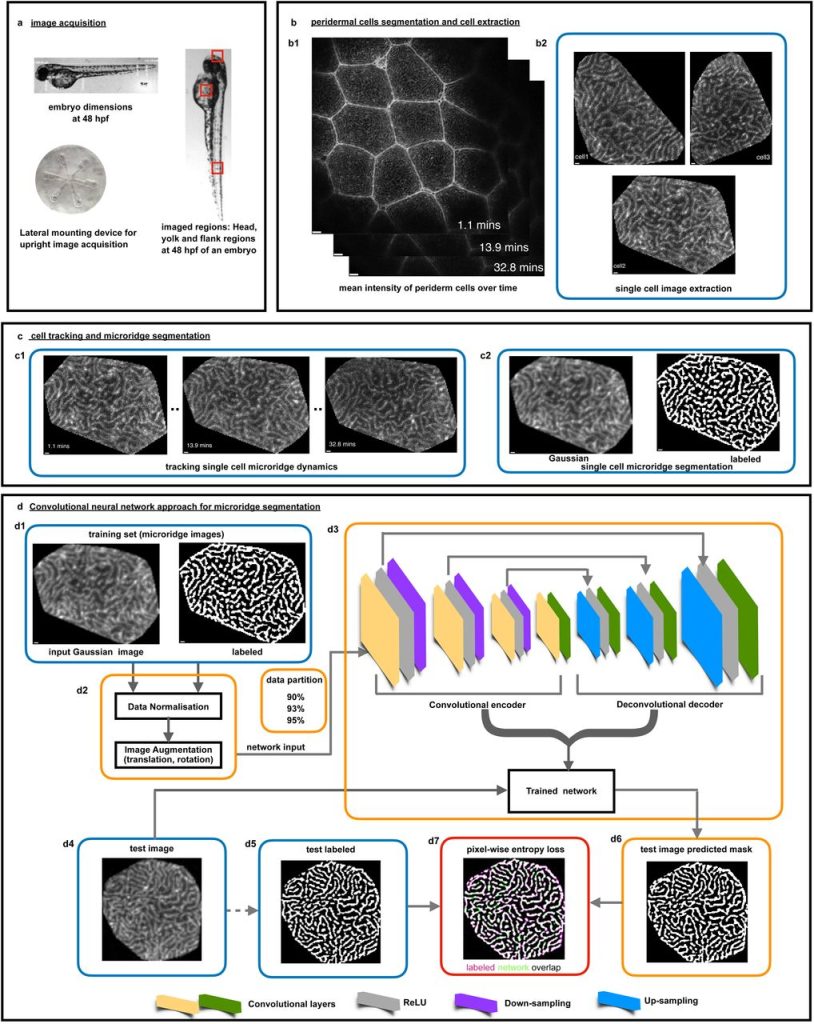
A deep learning framework for quantitative analysis of actin microridges. Rajasekaran Bhavna, Mahendra Sonawane
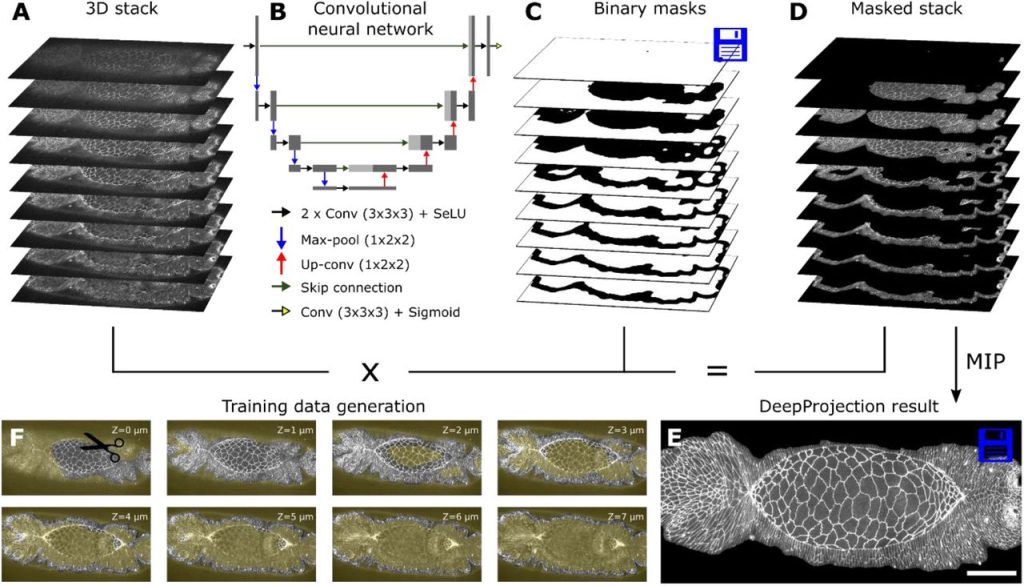
DeepProjection: Rapid and structure-specific projections of tissue sheets embedded in 3D microscopy stacks using deep learning. Daniel Haertter, Xiaolei Wang, Stephanie M. Fogerson, Nitya Ramkumar, Janice M. Crawford, Kenneth D. Poss, Stefano Di Talia, Daniel P. Kiehart, Christoph F. Schmidt
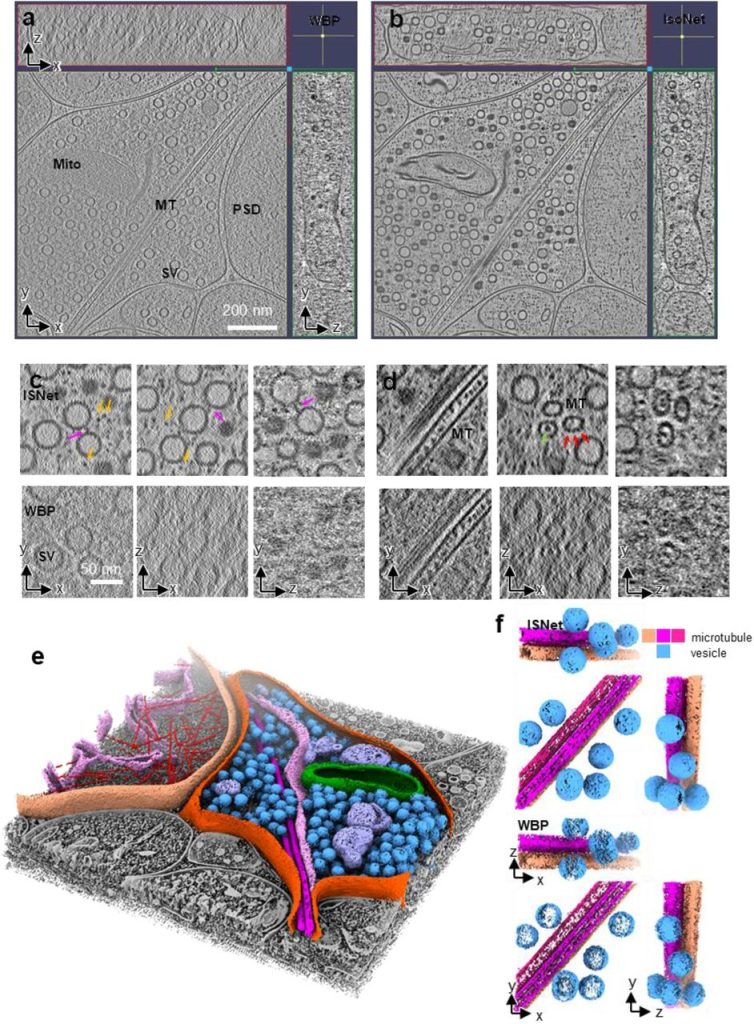
Isotropic Reconstruction of Electron Tomograms with Deep Learning. Yun-Tao Liu, Heng Zhang, Hui Wang, Chang-Lu Tao, Guo-Qiang Bi, Z. Hong Zhou
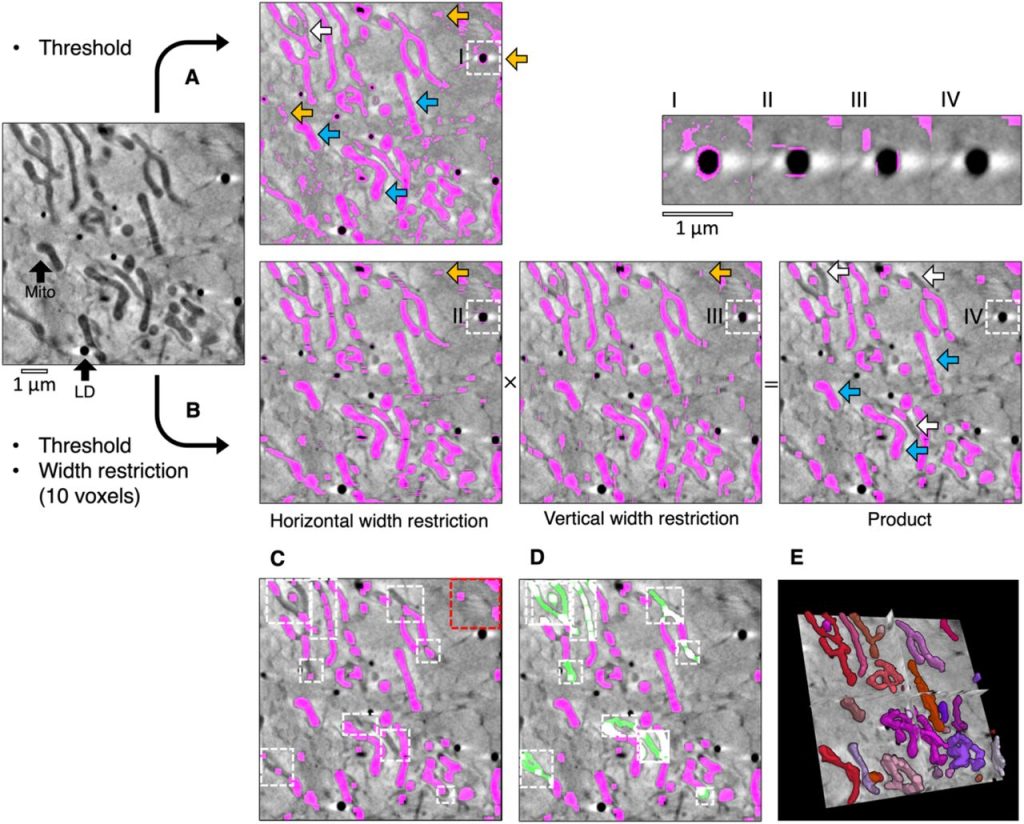
Contour, a semi-automated segmentation and quantitation tool for cryo-soft-X-ray tomography. Kamal L Nahas, João Ferreira Fernandes, Colin Crump, Stephen Graham, Maria Harkiolaki
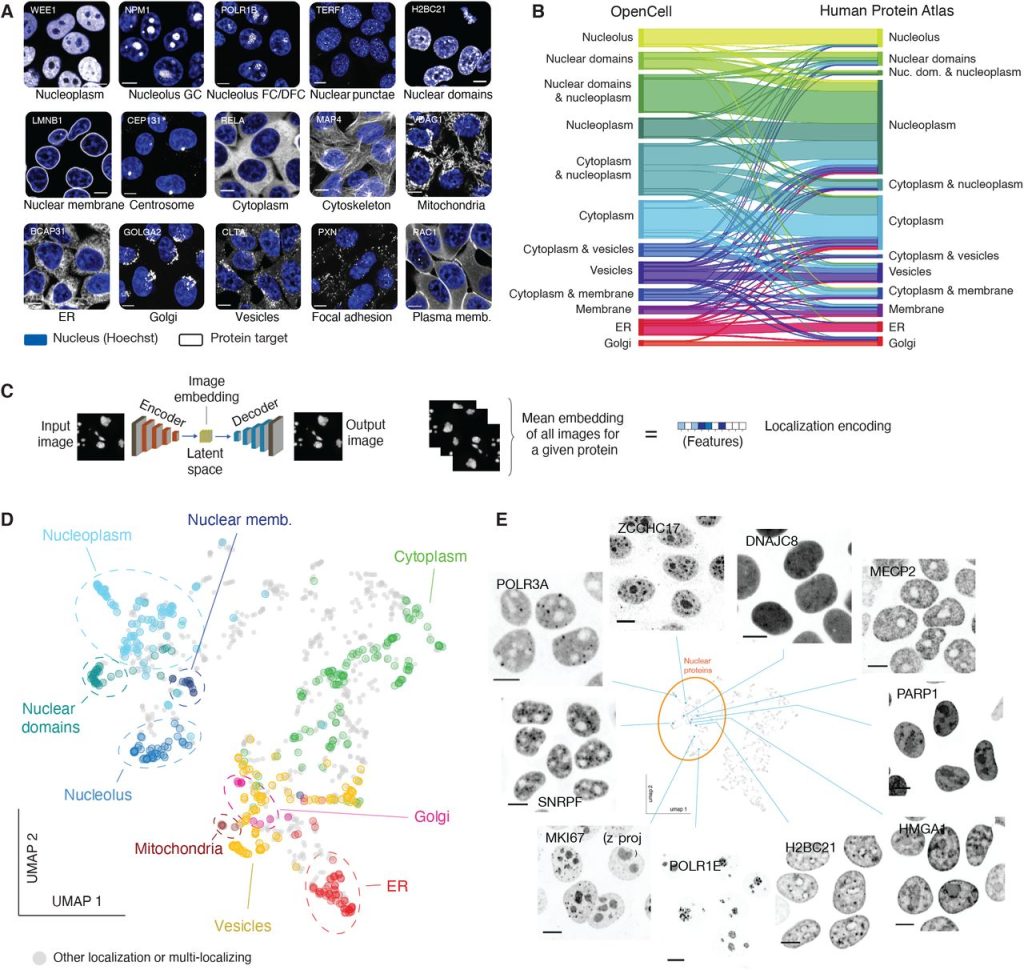
OpenCell: proteome-scale endogenous tagging enables the cartography of human cellular organization. Nathan H. Cho, Keith C. Cheveralls, Andreas-David Brunner, Kibeom Kim, André C. Michaelis, Preethi Raghavan, Hirofumi Kobayashi, Laura Savy, Jason Y. Li, Hera Canaj, James Y.S. Kim, Edna M. Stewart, Christian Gnann, Frank McCarthy, Joana P. Cabrera, Rachel M. Brunetti, Bryant B. Chhun, Greg Dingle, Marco Y. Hein, Bo Huang, Shalin B. Mehta, Jonathan S. Weissman, Rafael Gómez-Sjöberg, Daniel N. Itzhak, Loic A. Royer, Matthias Mann, Manuel D. Leonetti
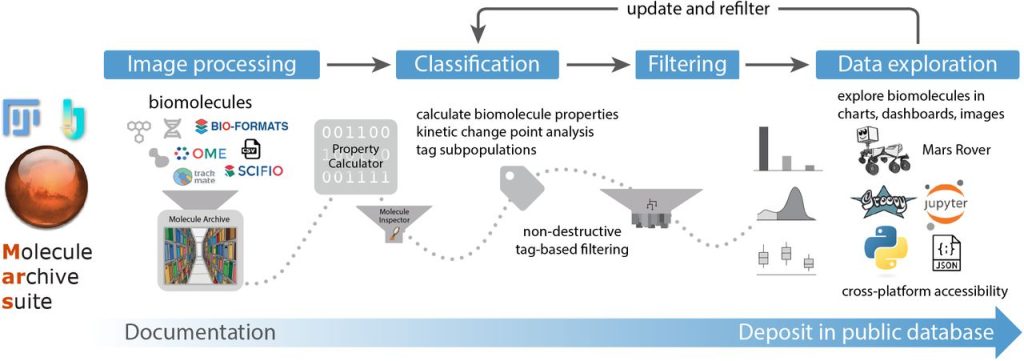
Mars, a molecule archive suite for reproducible analysis and reporting of single molecule properties from bioimages. Nadia M Huisjes, Thomas M Retzer, Matthias J Scherr, Rohit Agarwal, Barbara Safaric, Anita Minnen, Karl E Duderstadt
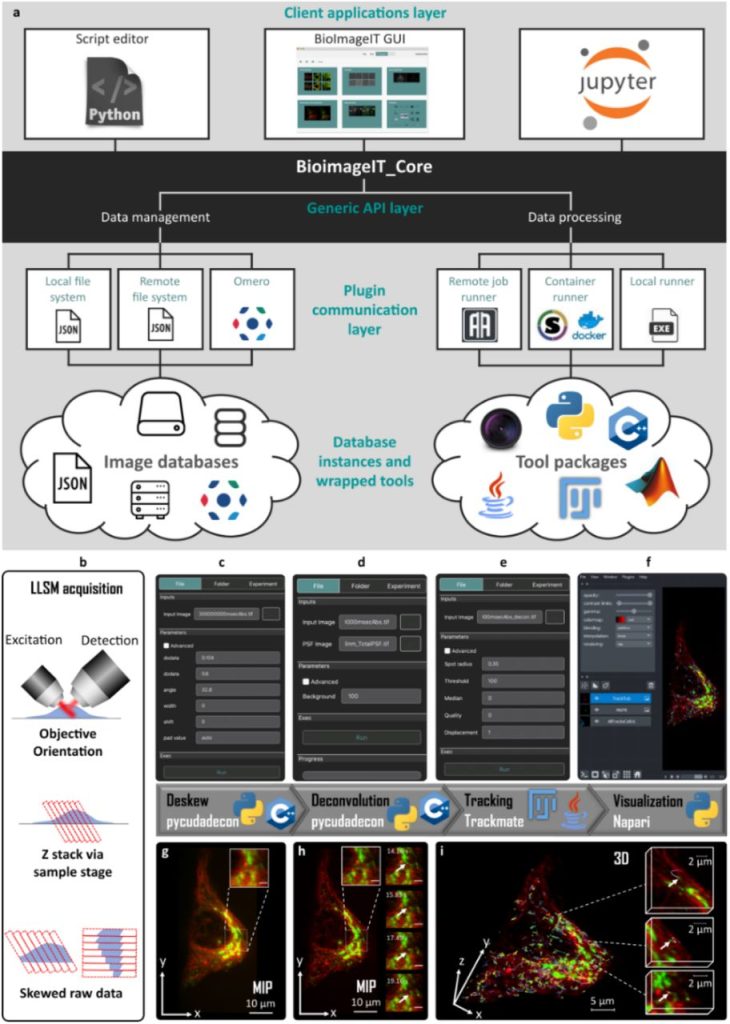
BioImageIT: Open-source framework for integration of image data-management with analysis. Sylvain Prigent, Cesar Augusto Valades-Cruz, Ludovic Leconte, Leo Maury, Jean Salamero, Charles Kervrann


 (No Ratings Yet)
(No Ratings Yet)