Microscopy preprints – Bioimage analysis tools
Posted by FocalPlane, on 28 January 2022
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools only.
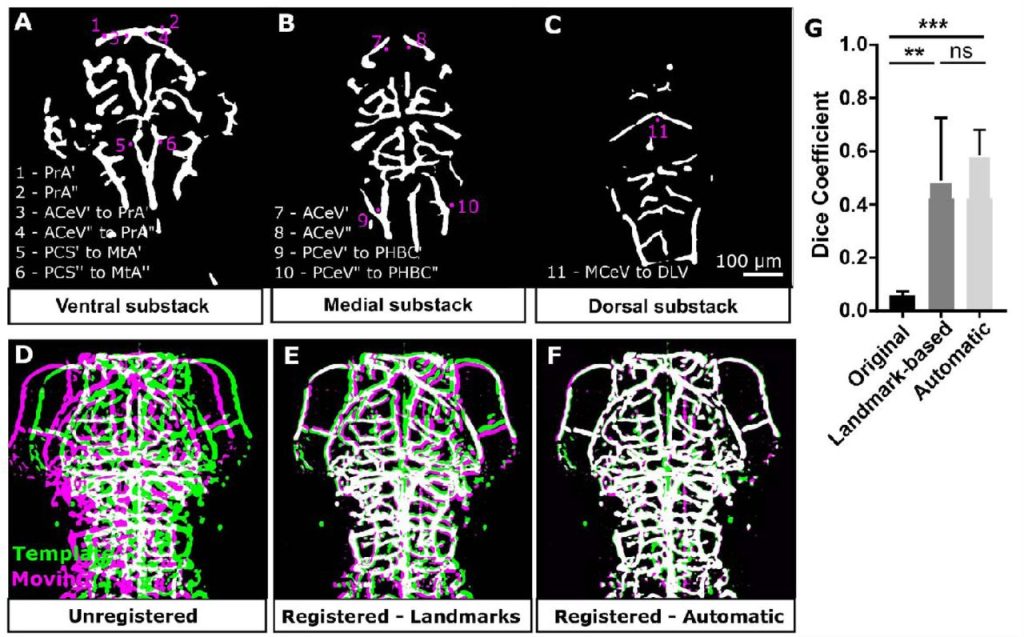
3D quantification of zebrafish cerebrovascular architecture by automated image analysis of light sheet fluorescence microscopy datasets. E. C. Kugler, J. Frost, V. Silva, K. Plant, K. Chhabria, T. J.A. Chico, P. A. Armitage
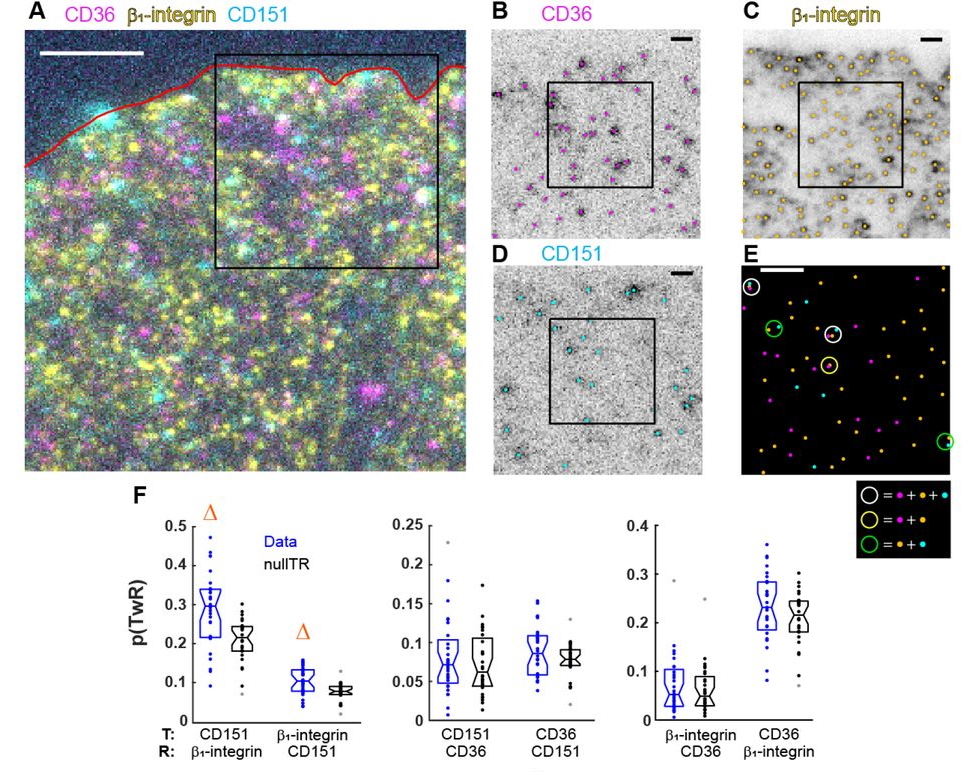
Analysis of conditional colocalization relationships and hierarchies from three-color microscopy images. Jesus Vega-Lugo, Bruno da Rocha-Azevedo, Aparajita Dasgupta, Nicolas Touret, Khuloud Jaqaman
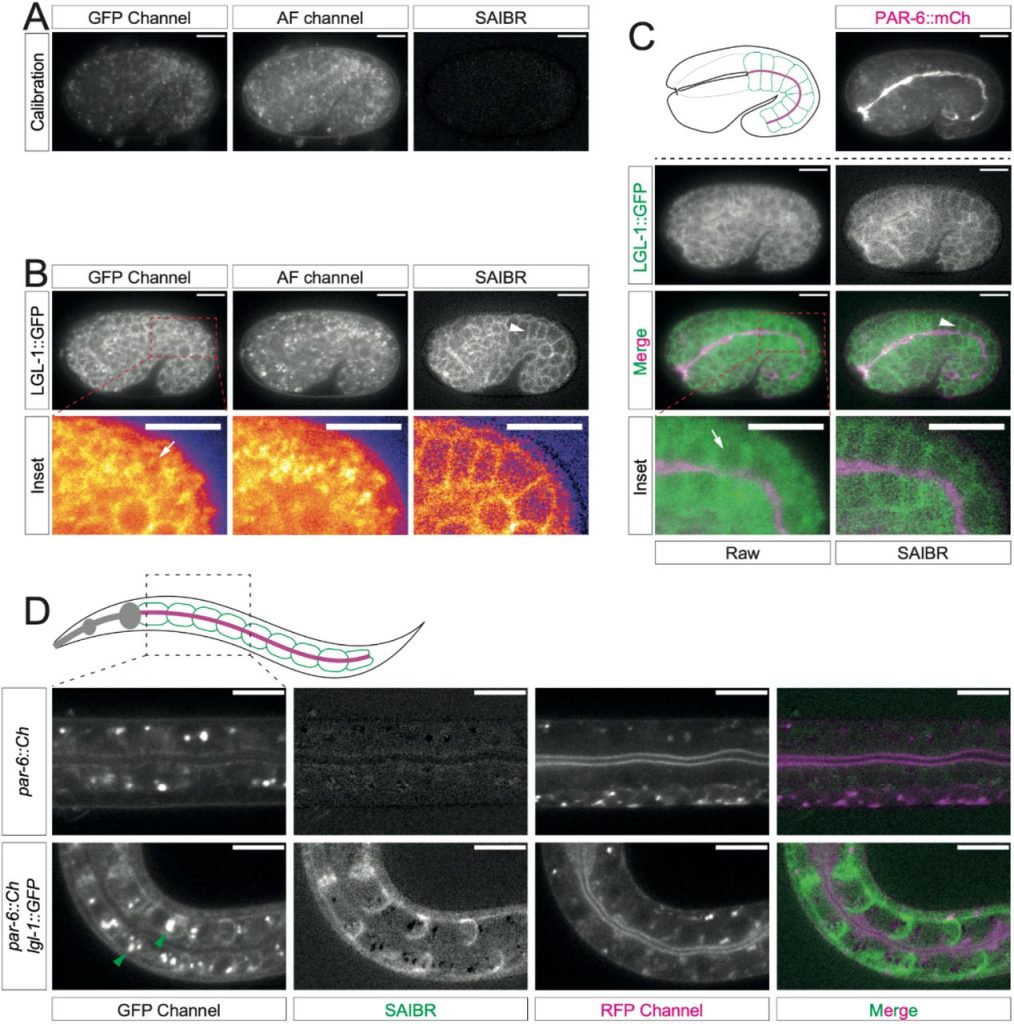
SAIBR: A simple, platform-independent method for spectral autofluorescence correction. Nelio T.L. Rodrigues, Tom Bland, Joana Borrego-Pinto, KangBo Ng, Nisha Hirani, Ying Gu, Sherman Foo, Nathan W Goehring
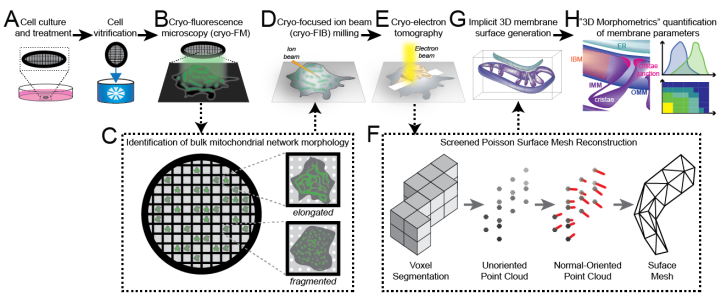
A surface morphometrics toolkit to quantify organellar membrane ultrastructure using cryo-electron tomography. Benjamin A Barad, Michaela Medina, Daniel Fuentes, R Luke Wiseman, Danielle A Grotjahn
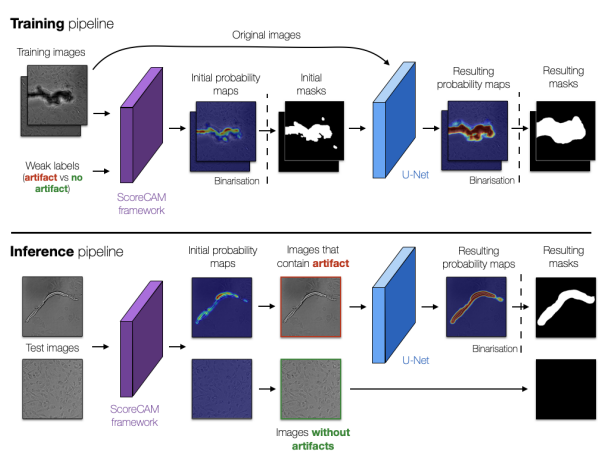
ArtSeg: Rapid Artifact Segmentation and Removal in Brightfield Cell Microscopy Images. Mohammed A. S. Ali, Kaspar Hollo, Tõnis Laasfled, Jane Torp, Maris-Johanna Tahk, Ago Rinken, Kaupo Palo, Leopold Parts, Dmytro Fishman
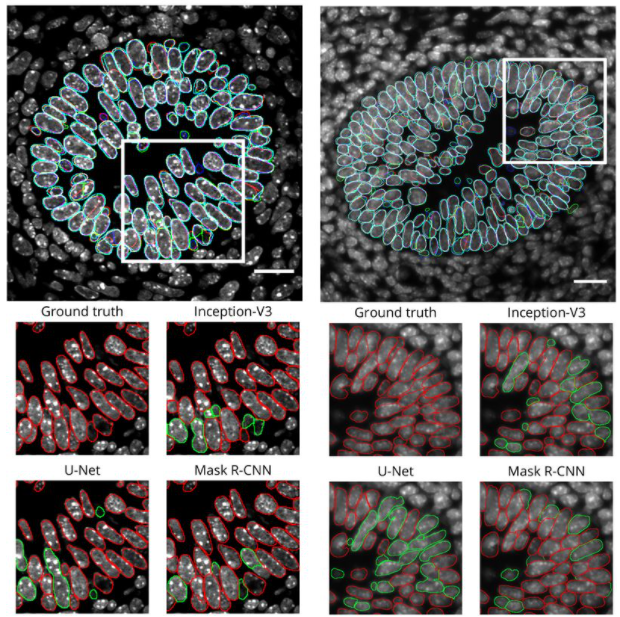
Deep learning tools and modeling to estimate the temporal expression of cell cycle proteins from 2D still images. Thierry Pécot, Maria C. Cuitiño, Roger H. Johnson, Cynthia Timmers, Gustavo Leone
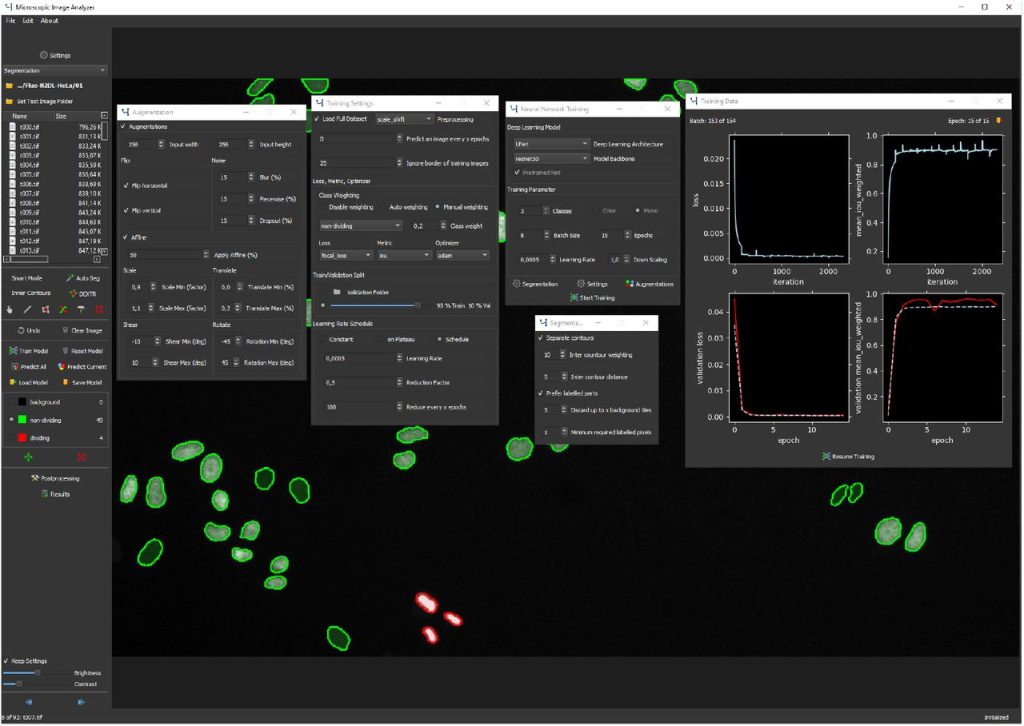
MIA: An Open Source Standalone Deep Learning Application for Microscopic Image Analysis. Nils Körber
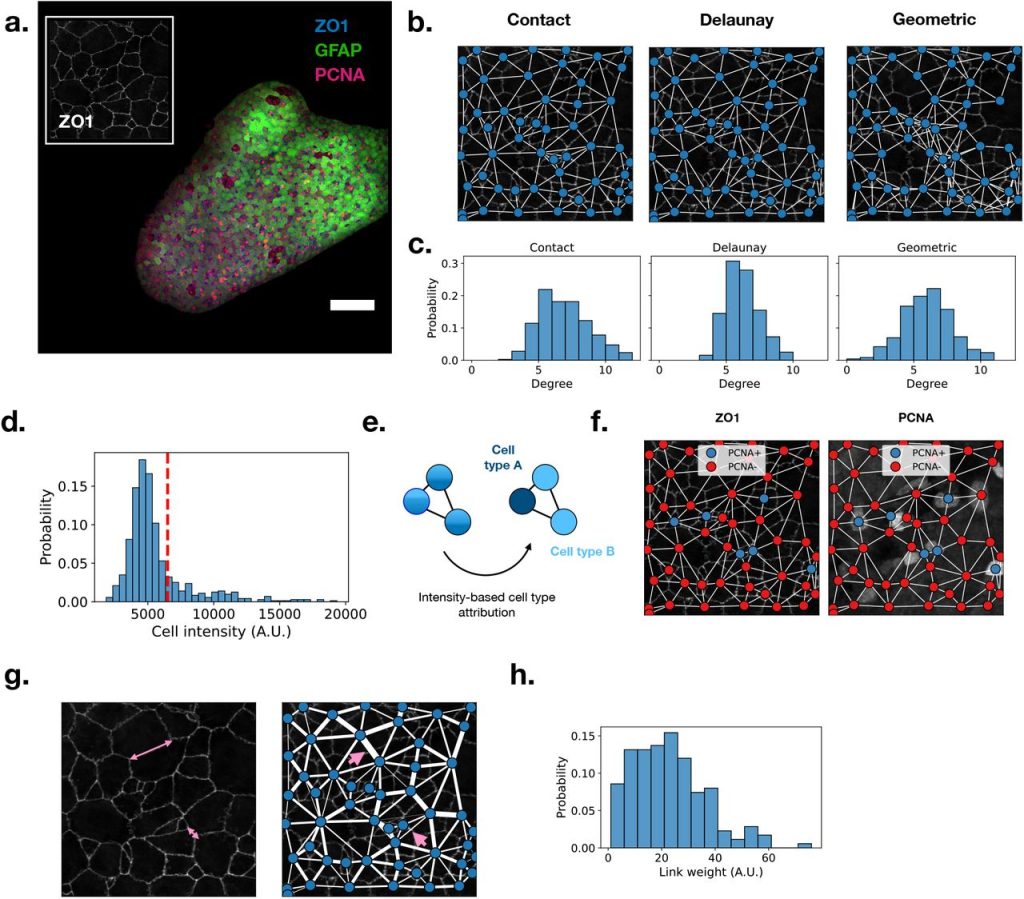
Griottes: a generalist tool for network generation from segmented tissue images. Gustave Ronteix, Valentin Bonnet, Sebastien Sart, Jeremie Sobel, Elric Esposito, Charles N. Baroud
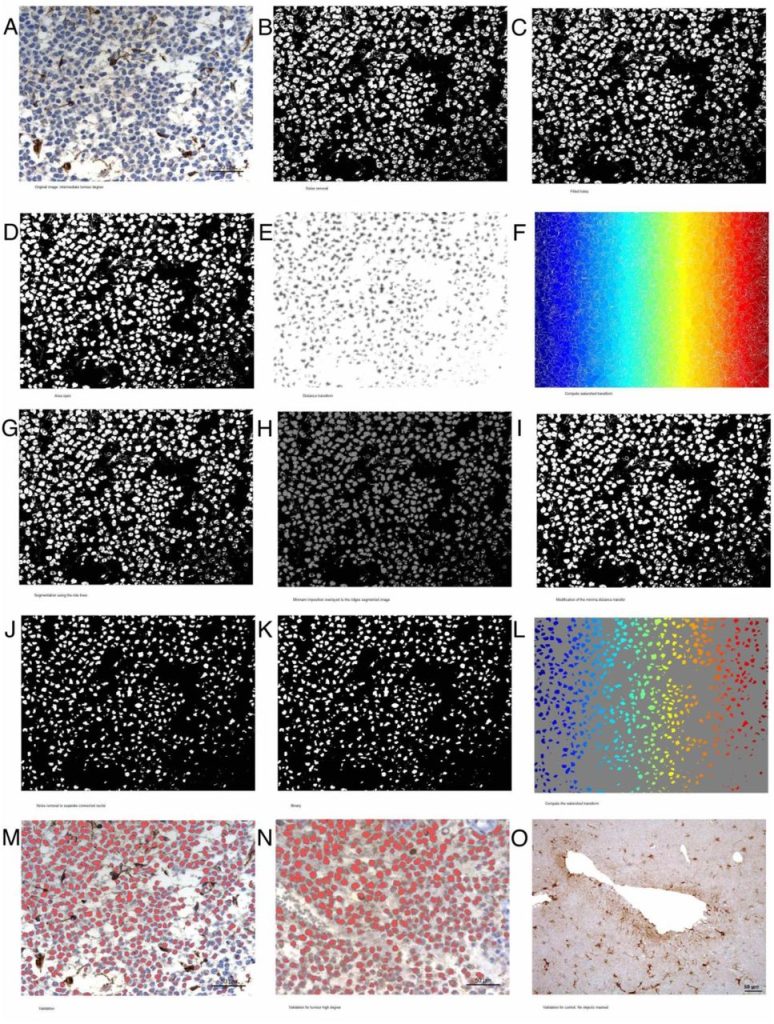
Validated method for automated glioma diagnosis from GFAP immunohistological images: a complete pipeline. A. Campo, F. Fernández-Flores, M. Pumarola
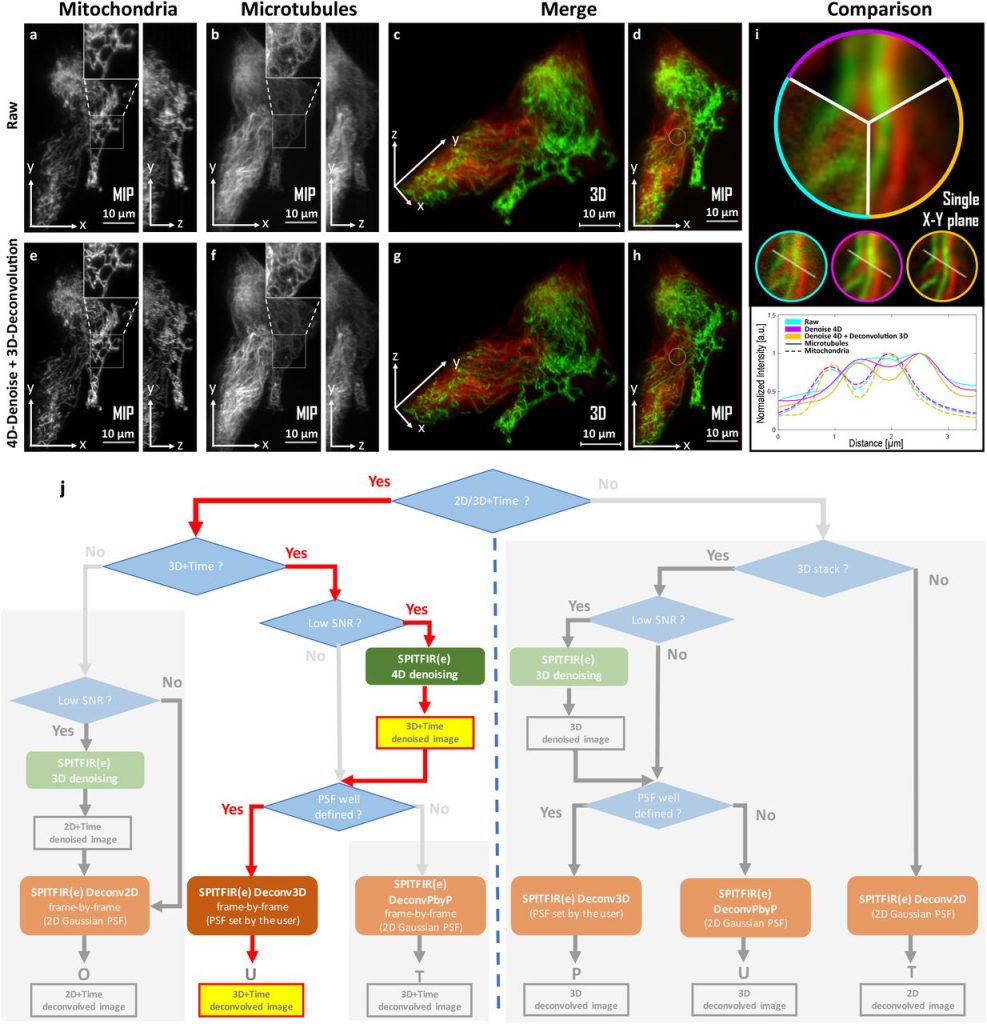
SPITFIR(e): A supermaneuverable algorithm for restoring 2D-3D fluorescence images and videos, and background subtraction. Sylvain Prigent, Hoai-Nam Nguyen, Ludovic Leconte, Cesar Augusto Valades-Cruz, Bassam Hajj, Jean Salamero, Charles Kervrann
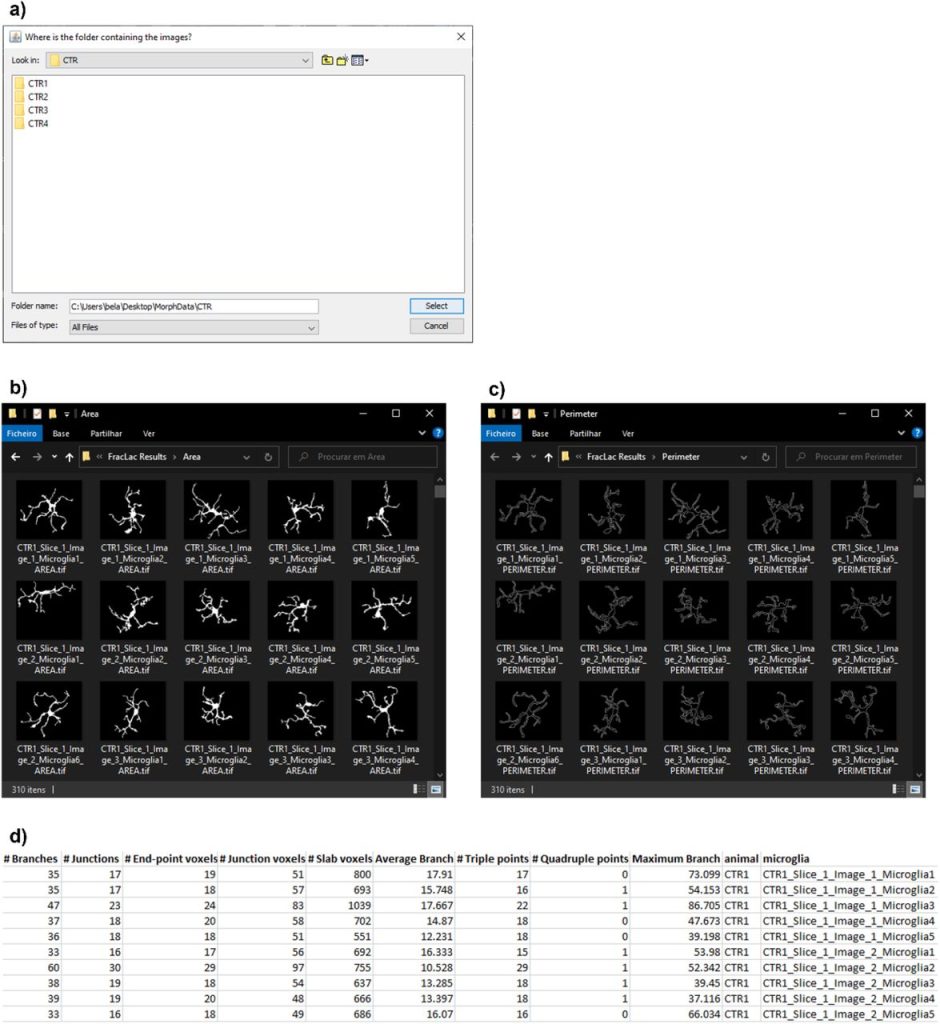
MorphData: Automating the data extraction process of morphological features of microglial cells in ImageJ. Ana Bela Campos, Sara Duarte-Silva, António Francisco Ambrósio, Patrícia Maciel, Bruno Fernandes
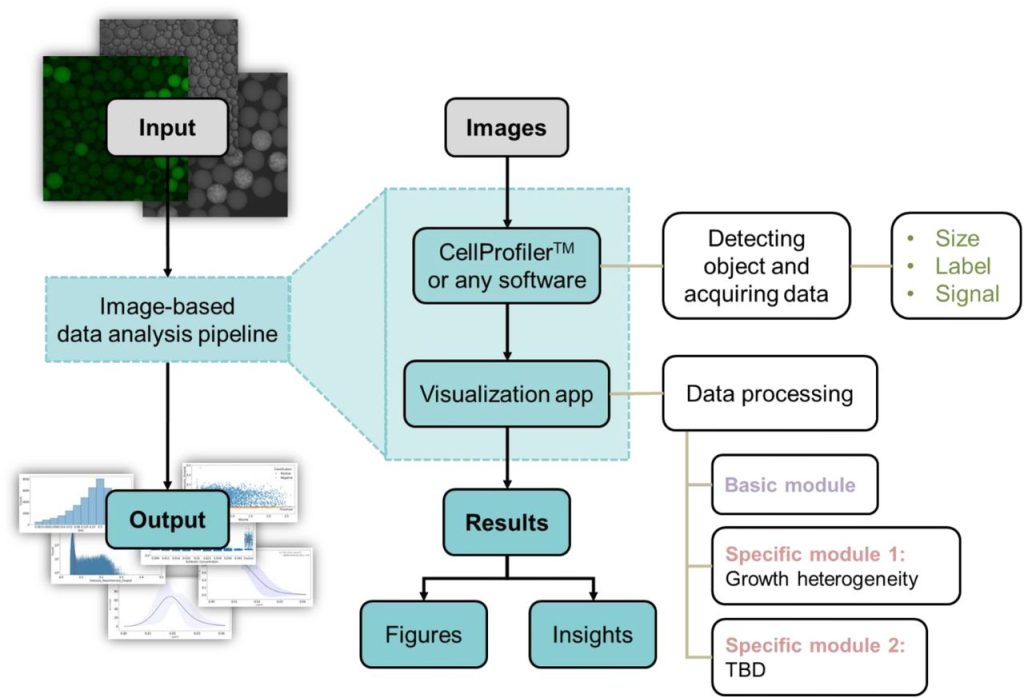
EasyFlow: User-friendly Workflow for Image-based Droplet Analysis with Multipurpose Modules. Immanuel Sanka, Simona Bartkova, Pille Pata, Karol Makuch, Olli-Pekka Smolander, Ott Scheler
Scellseg: a style-aware cell instance segmentation tool with pre-training and contrastive fine-tuning. Dejin Xun, Deheng Chen, Yitian Zhou, Volker M. Lauschke, Rui Wang, Yi Wang
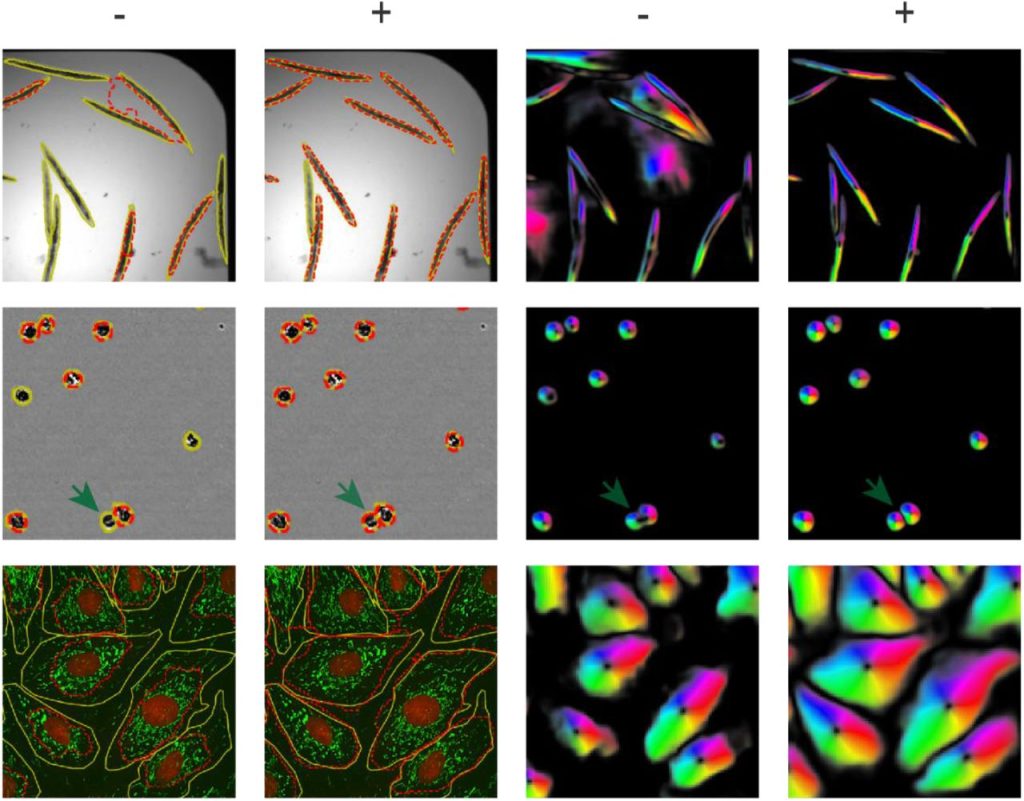
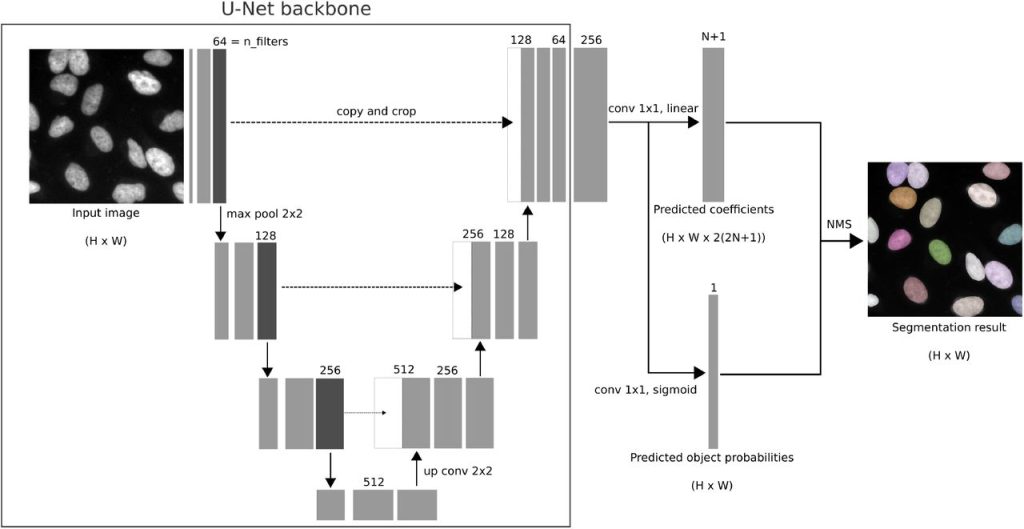
Fully Automatic Cell Segmentation with Fourier Descriptors. Dominik Hirling, Peter Horvath
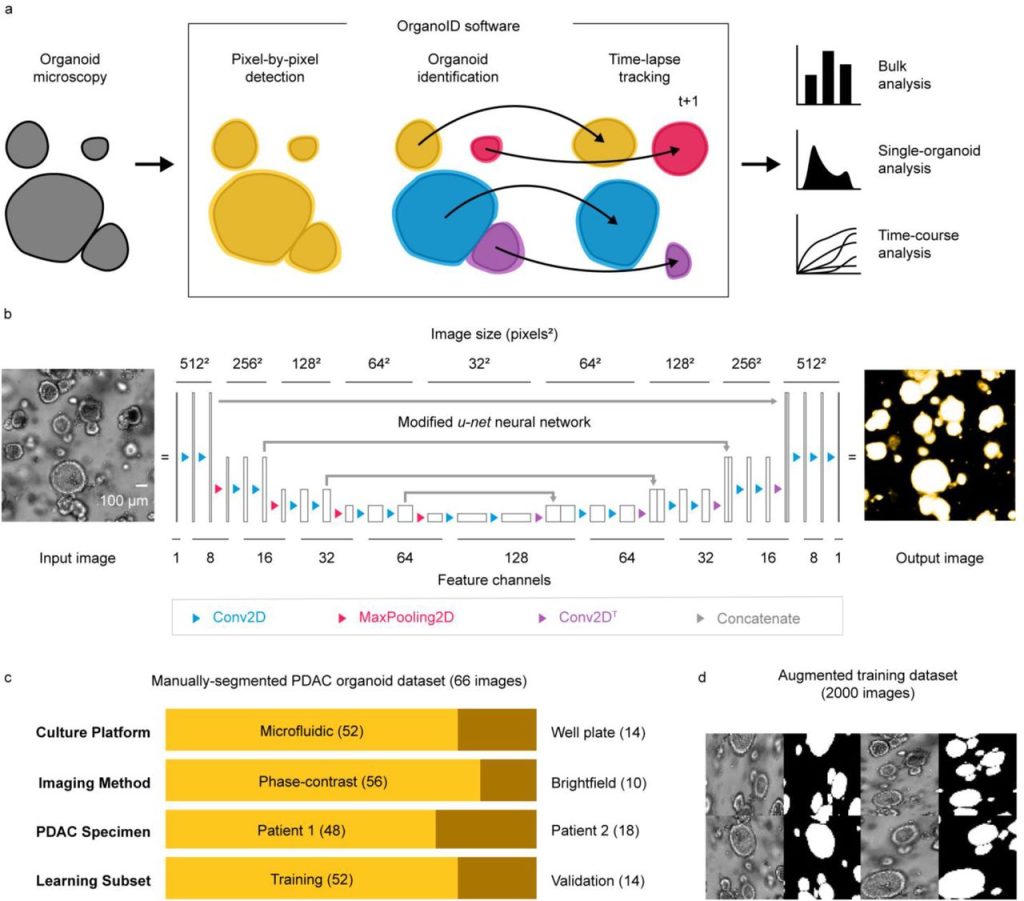
OrganoID: a versatile deep learning platform for organoid image analysis. Jonathan Matthews, Brooke Schuster, Sara Saheb Kashaf, Ping Liu, Mustafa Bilgic, Andrey Rzhetsky, Savaş Tay
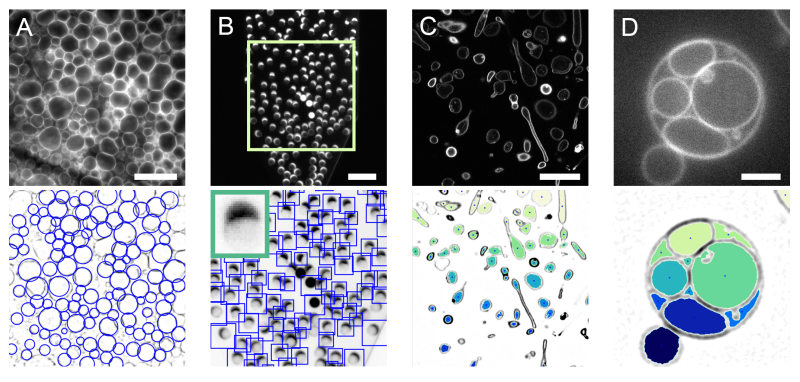
DisGUVery: a versatile open-source software for high-throughput image analysis of Giant Unilamellar Vesicles. Lennard van Buren, Gijsje H. Koenderink, Cristina Martinez-Torres
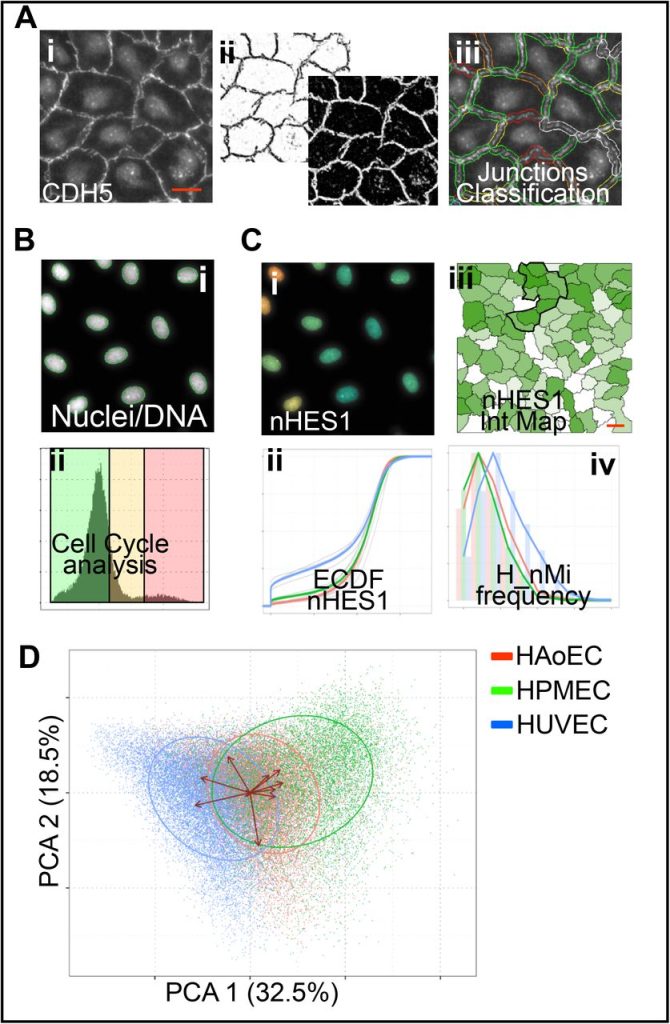
High content Image Analysis to study phenotypic heterogeneity in endothelial cell monolayers. Francois Chesnais, Jonas Hue, Errin Roy, Marco Branco, Ruby Stokes, Aize Pellon, Juliette Le Caillec, Eyad Elbahtety, Matteo Battilocchi, Davide Danovi, Lorenzo Veschini
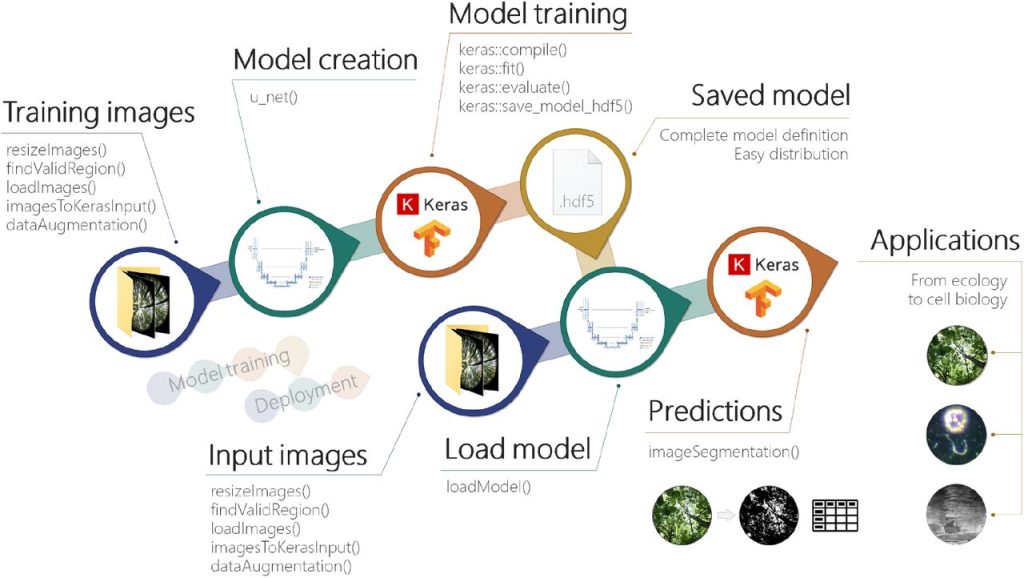
imageseg: an R package for deep learning-based image segmentation. Jürgen Niedballa, Jan Axtner, Timm Fabian Döbert, Andrew Tilker, An Nguyen, Seth T. Wong, Christian Fiderer, Marco Heurich, Andreas Wilting
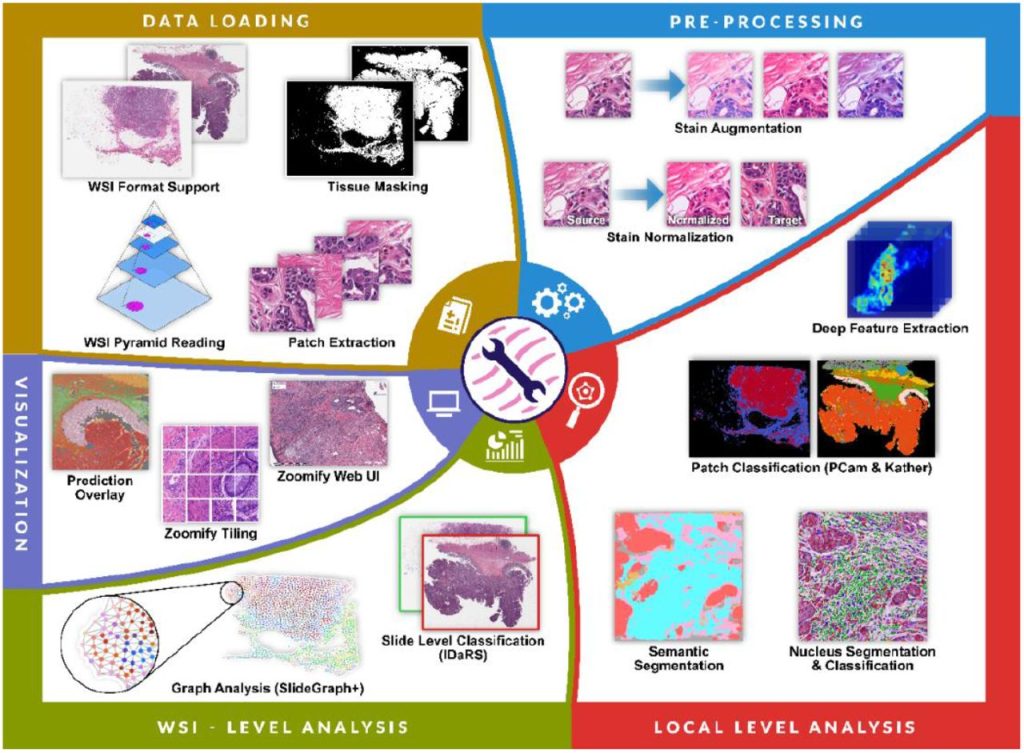
TIAToolbox: An End-to-End Toolbox for Advanced Tissue Image Analytics. Johnathan Pocock, Simon Graham, Quoc Dang Vu, Mostafa Jahanifar, Srijay Deshpande, Giorgos Hadjigeorghiou, Adam Shephard, Raja Muhammad Saad Bashir, Mohsin Bilal, Wenqi Lu, David Epstein, Fayyaz Minhas, Nasir M. Rajpoot, Shan E Ahmed Raza


 (4 votes, average: 1.00 out of 1)
(4 votes, average: 1.00 out of 1)