Microscopy preprints – Bioimage analysis tools
Posted by FocalPlane, on 11 March 2022
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools only.
Cloud-enabled Fiji for reproducible, integrated and modular image processing. Ling-Hong Hung, Evan Straw, Shishir Reddy, Robert Schmitz, Zachary Colburn, Ka Yee Yeung
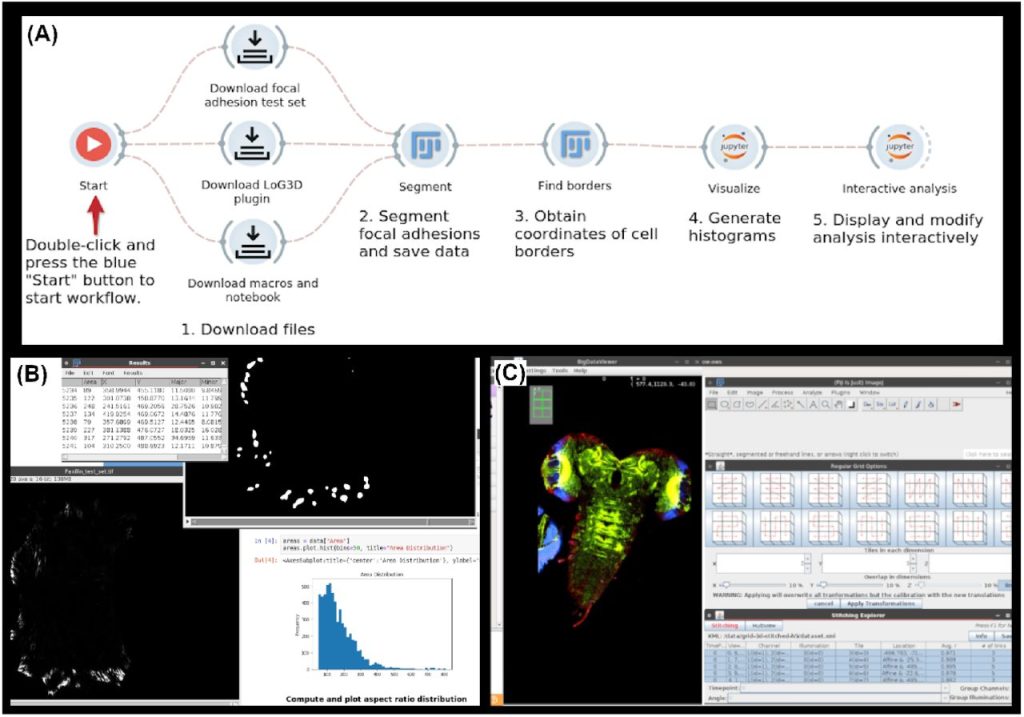
A pipeline to track unlabeled cells in wide migration chambers using pseudofluorescence. Antonello Paola, Marcus Thelen, Rolf Krause, Pizzagalli Diego Ulisse
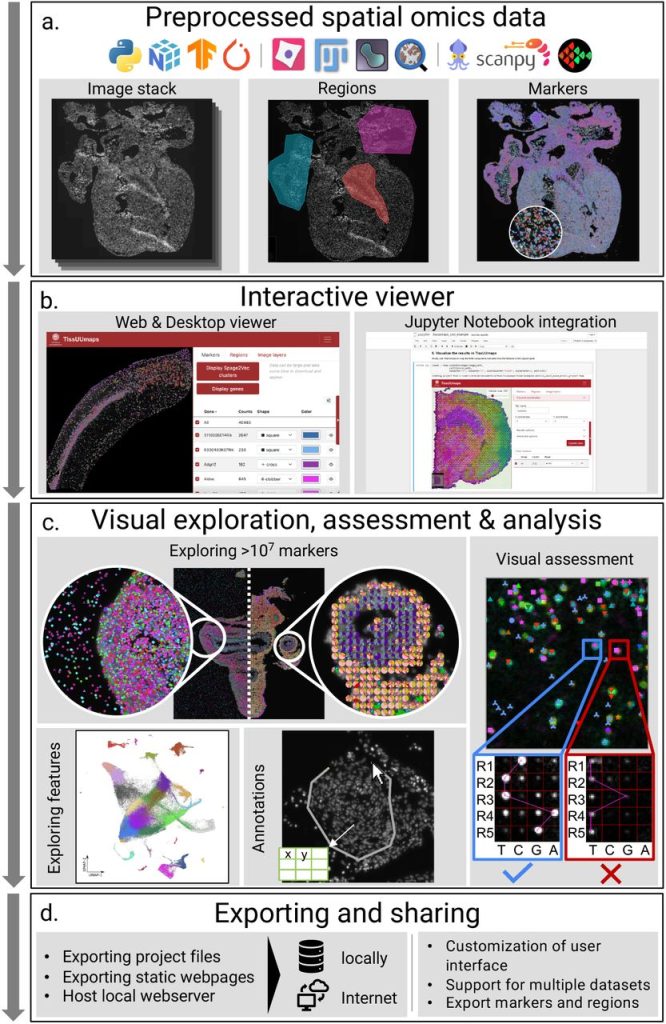
TissUUmaps 3: Interactive visualization and quality assessment of large-scale spatial omics data. Nicolas Pielawski, Axel Andersson, Christophe Avenel, Andrea Behanova, Eduard Chelebian, Anna Klemm, Fredrik Nysjö, Leslie Solorzano, Carolina Wählby
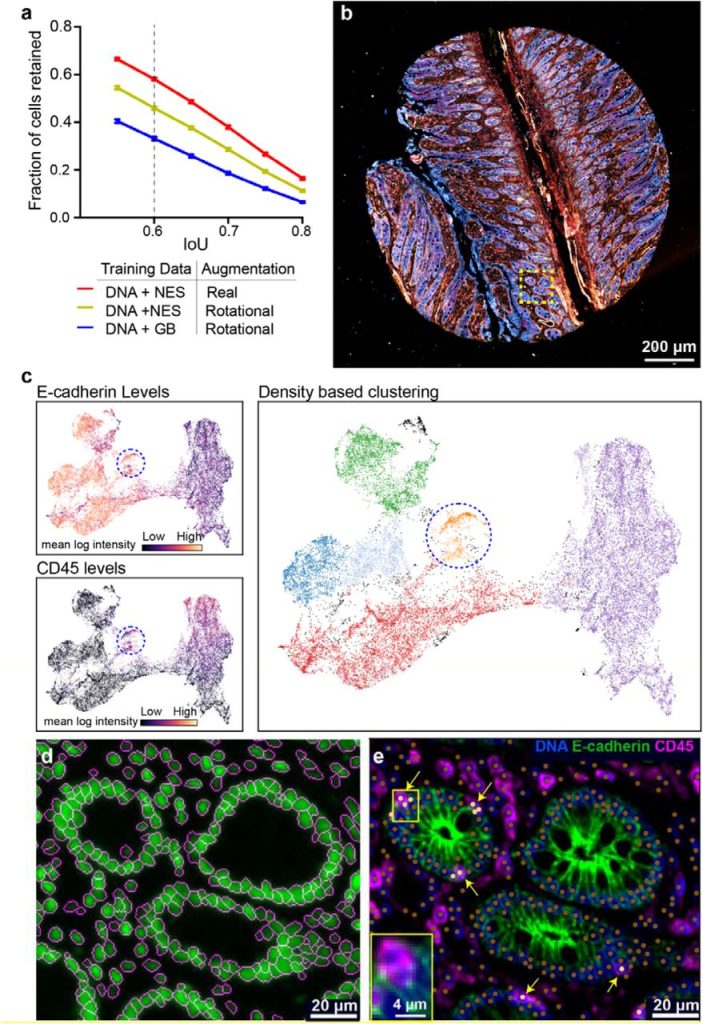
UnMICST: Deep learning with real augmentation for robust segmentation of highly multiplexed images of human tissues. Clarence Yapp, Edward Novikov, Won-Dong Jang, Tuulia Vallius, Yu-An Chen, Marcelo Cicconet, Zoltan Maliga, Connor A. Jacobson, Donglai Wei, Sandro Santagata, Hanspeter Pfister, Peter K. Sorger
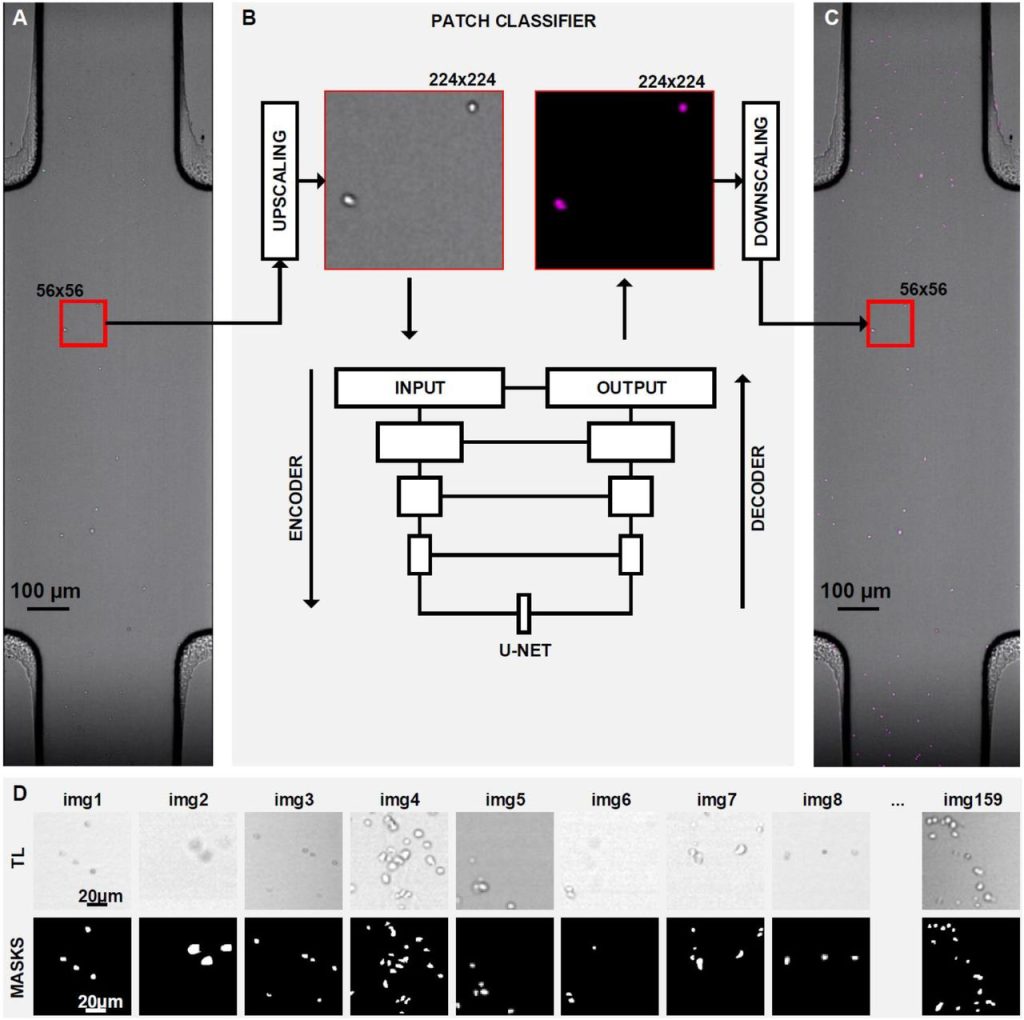
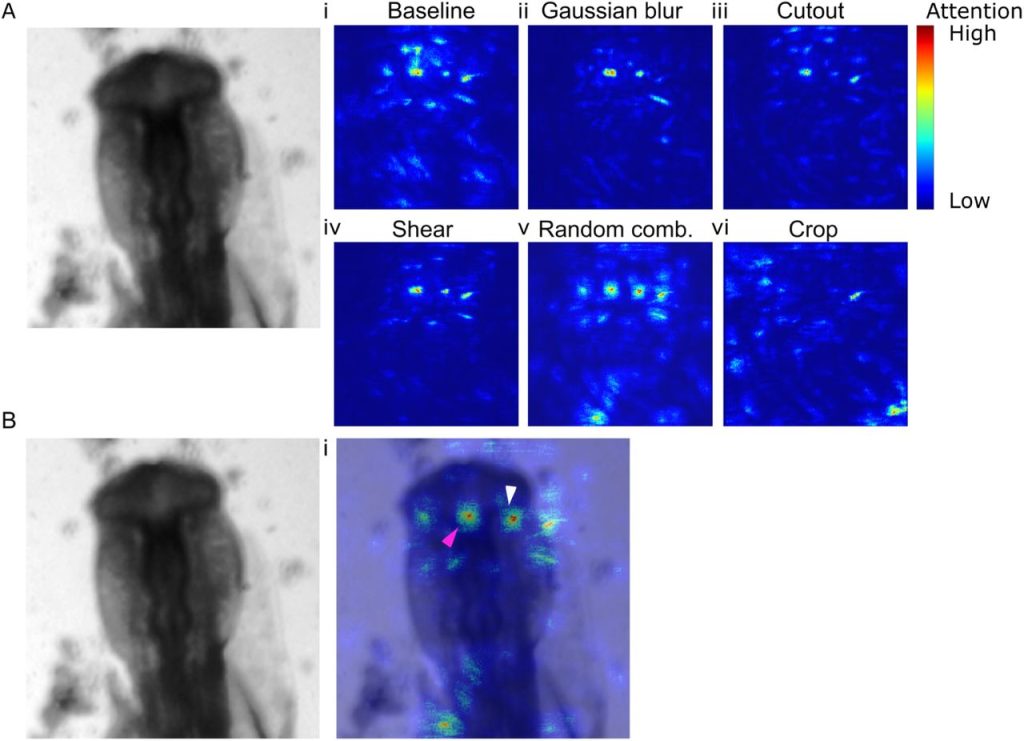
Bespoke data augmentation and network construction enable image classification on small microscopy datasets. Ian Groves, Jacob Holmshaw, David Furley, Benjamin D. Evans, Marysia Placzek, Alexander G. Fletcher
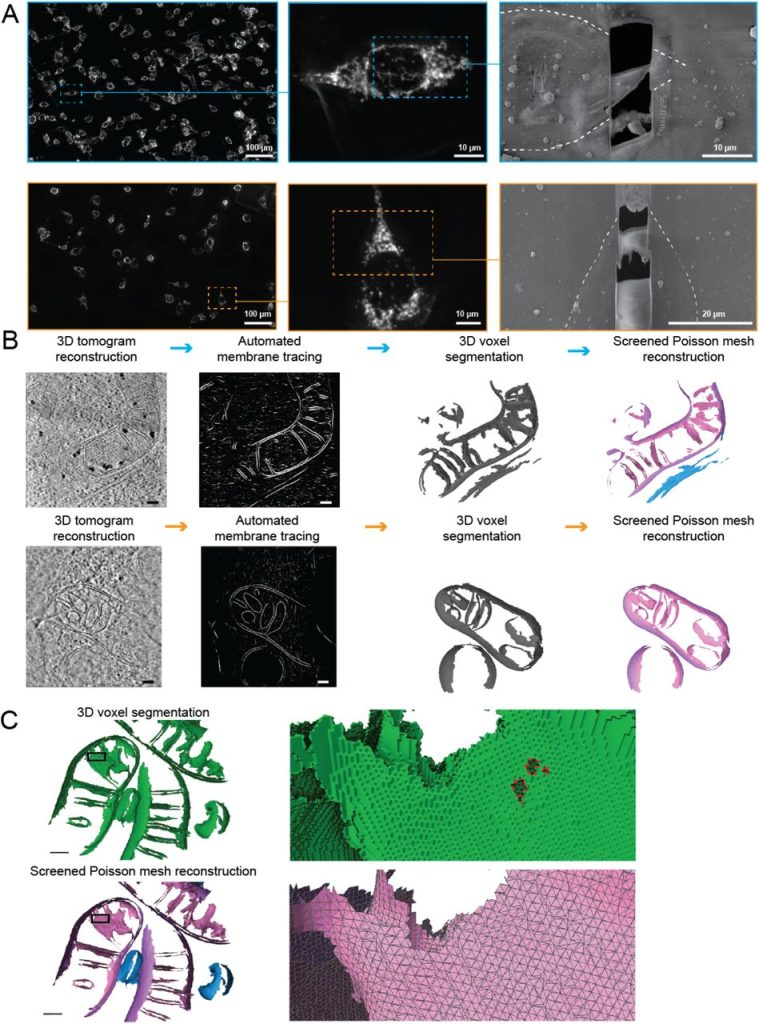
A surface morphometrics toolkit to quantify organellar membrane ultrastructure using cryo-electron tomography. Benjamin A Barad, Michaela Medina, Daniel Fuentes, R Luke Wiseman, Danielle A Grotjahn
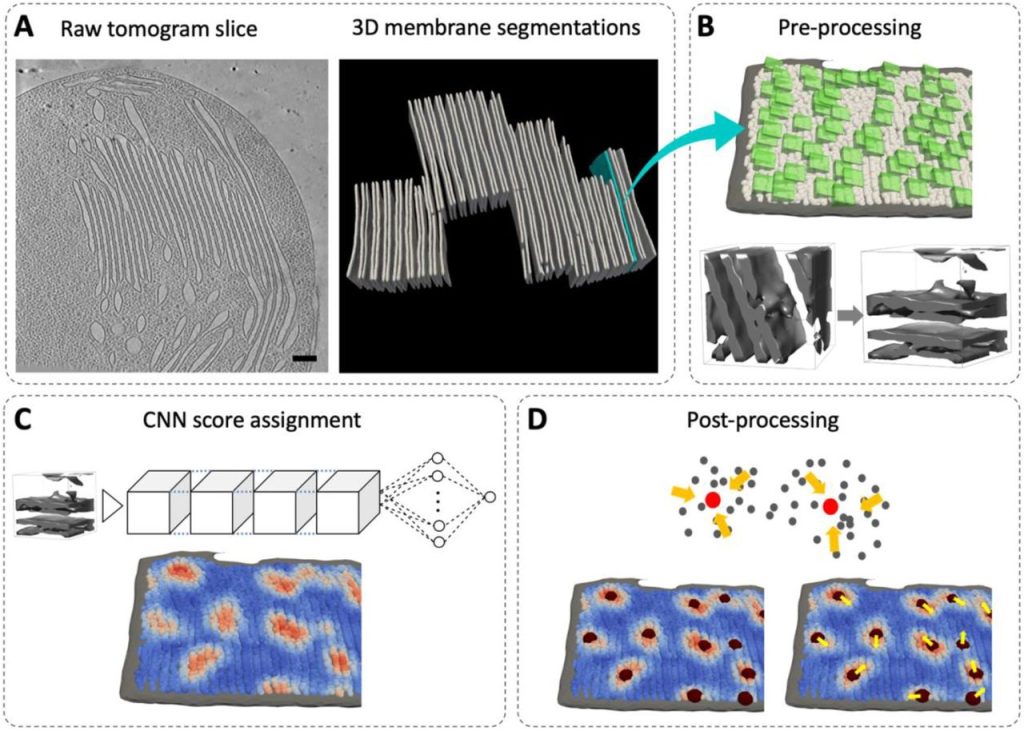
MemBrain: A Deep Learning-aided Pipeline for Automated Detection of Membrane Proteins in Cryo-electron Tomograms. Lorenz Lamm, Ricardo D. Righetto, Wojciech Wietrzynski, Matthias Pöge, Antonio Martinez-Sanchez, Tingying Peng, Benjamin D. Engel
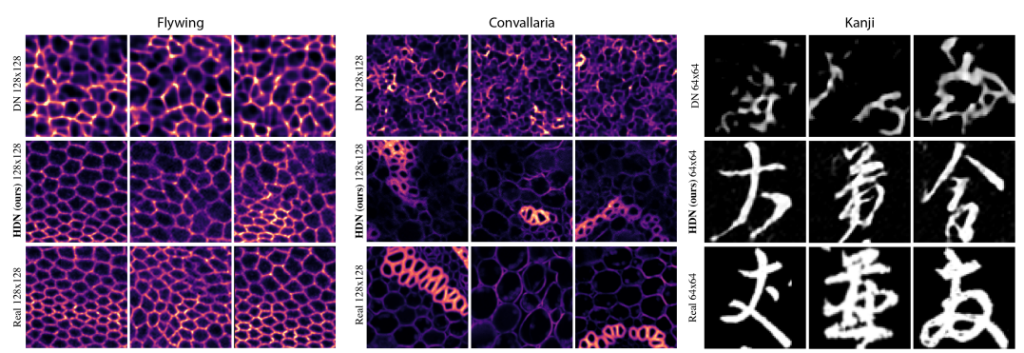
Interpretable Unsupervised Diversity Denoising and Artefact Removal. Mangal Prakash, Mauricio Delbracio, Peyman Milanfar, Florian Jug
Machine learning meets classical computer vision for accurate cell identification. Elham Karimi, Morteza Rezanejad, Benoit Fiset, Lucas Perus, Sheri A.C. McDowell, Azadeh Arabzadeh, Gaspard Beugnot, Peter Siegel, Marie-Christine Guiot, Daniela F. Quail, Kaleem Siddiqi, Logan A. Walsh
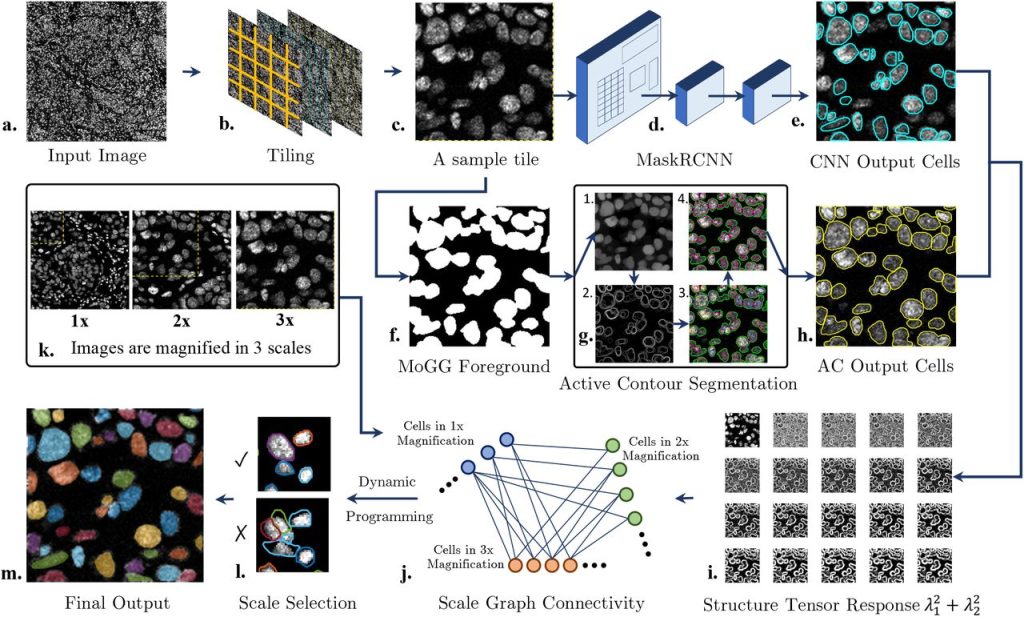
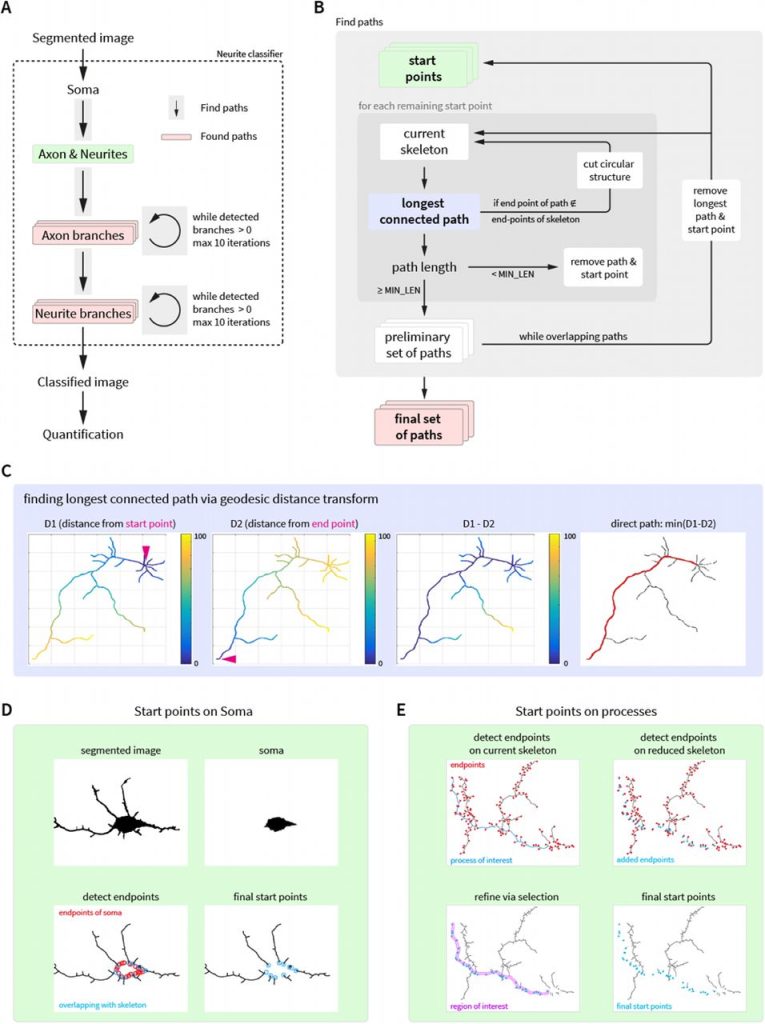
Automated Analysis of Neuronal Morphology through an Unsupervised Classification Model of Neurites. Amin Zehtabian, Joachim Fuchs, Britta Eickholt, Helge Ewers
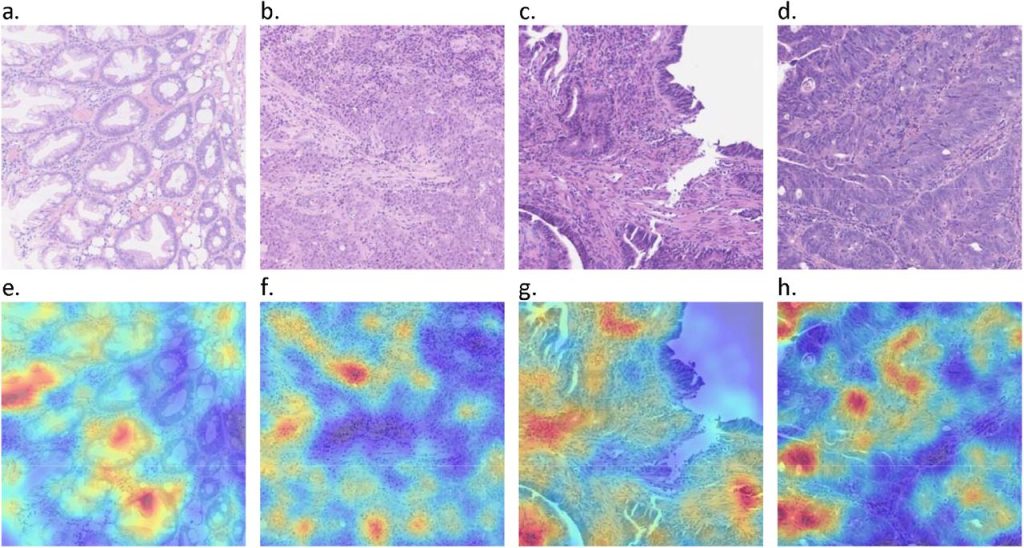
AI based pre-screening of large bowel cancer via weakly supervised learning of colorectal biopsy histology images. Mohsin Bilal, Yee Wah Tsang, Mahmoud Ali, Simon Graham, Emily Hero, Noorul Wahab, Katherine Dodd, Harvir Sahota, Wenqi Lu, Mostafa Jahanifar, Andrew Robinson, Ayesha Azam, Ksenija Benes, Mohammed Nimir, Abhir Bhalerao, Hesham Eldaly, Shan E Ahmed Raza, Kishore Gopalakrishnan, Fayyaz Minhas, David Snead, Nasir Rajpoot
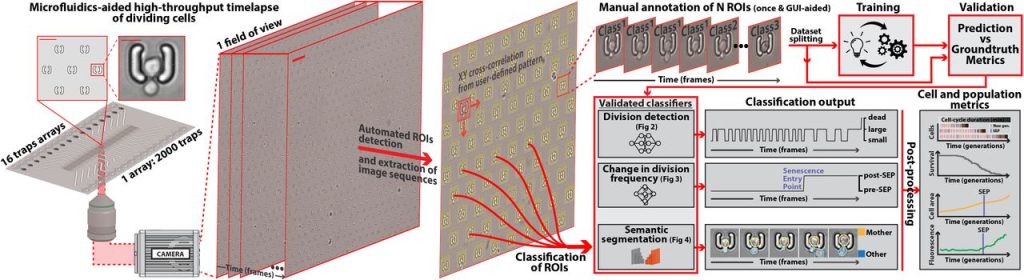
DetecDiv, a deep-learning platform for automated cell division tracking and replicative lifespan analysis. Théo Aspert, Didier Hentsch, Gilles Charvin
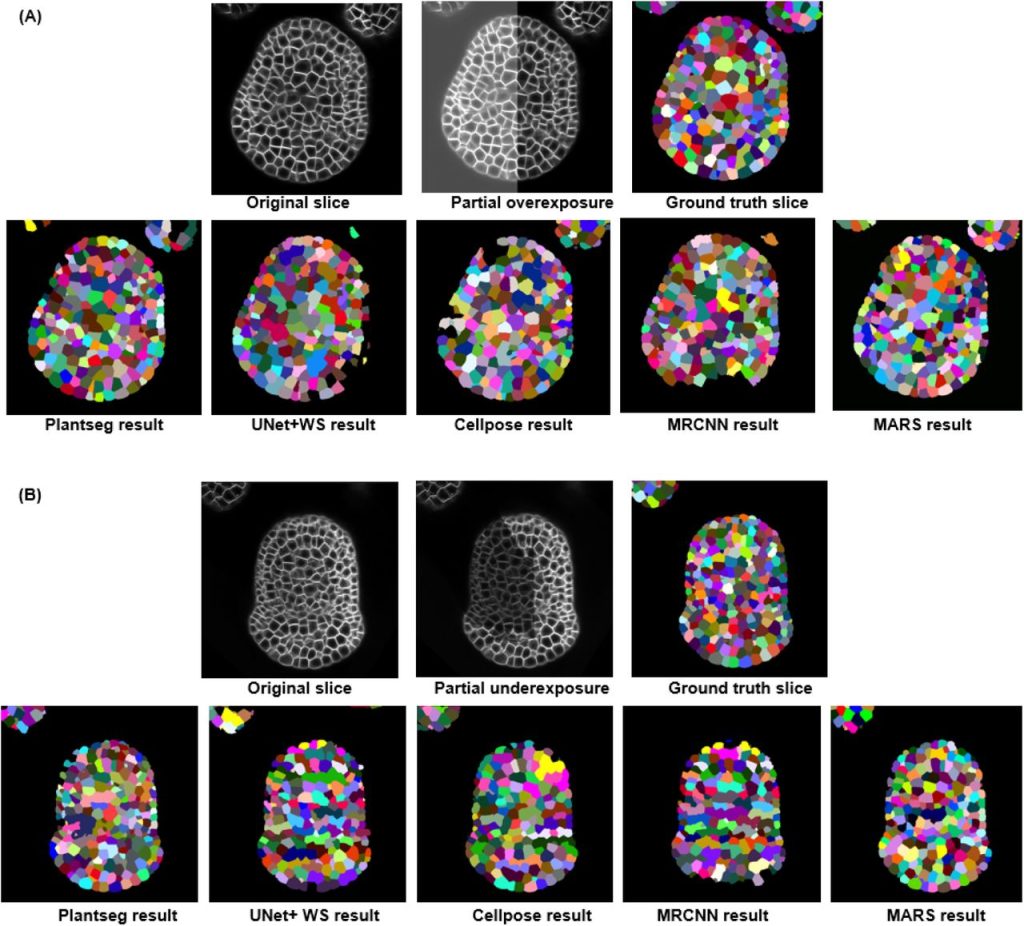
Benchmarking of deep learning algorithms for 3D instance segmentation of confocal image datasets. Anuradha Kar, Manuel Petit, Yassin Refahi, Guillaume Cerutt, Christophe Godin, Jan Traas


 (No Ratings Yet)
(No Ratings Yet)