Cell Worlds: meet the microscopists
Posted by FocalPlane, on 14 July 2022
February, 2022 saw the world premiere of the Cell Worlds documentary and exhibition. Cell Worlds was created by co-founders Renaud Pourpre and Terence Saulnier to bring microscopy out of the lab and display it in new ways to new audiences. Alongside our sister site, the Node, we wanted to find out more about the people behind this fantastic outreach project. Here, we focus on some of the microscopists who provided the images and movies. You can find an interview about the story behind the project with Renaud and Terence, the co-founders of Cell Worlds, on the Node.
Hoang Anh Le
Research: My research during my PhD focused on the regulation of cancer cell migration and metastasis. A large part of my PhD revolves around a protein of unknown function called CYRI-A (or Fam49A). I utilise many different microscopy techniques, including Airyscan super resolution microscopy and long term timelapse, on which many of my videos featured on the Cell World documentary were filmed.

How did you become involved with the project? I never expected to be contacted by anyone about my images to be honest. During my third year, I started to develop a habit of sharing some of my microscopy images, mostly from failed experiments, on Twitter. One of the most viral videos was a timelapse of two cell lines, one invasive and one non-invasive. Renaud and Terence must have seen this video and contacted my PhD supervisor, Laura Machesky, during the second lockdown to ask if I and Laura wanted to join their project. And of course, we were very surprised but thrilled and accepted their offer, and the rest is history!
What do you think about Cell Worlds? The project deserves all the praise and success that it has gathered. I showed the documentary to my family back home and they are mesmerised, and they don’t even speak French or English. That just shows how impactful this project is, the stunning visual effects complimented by the incredible music and so even people who aren’t in the field can still appreciate the beauty of science. Renaud and Terence and their team have done an incredible job turning this into reality.
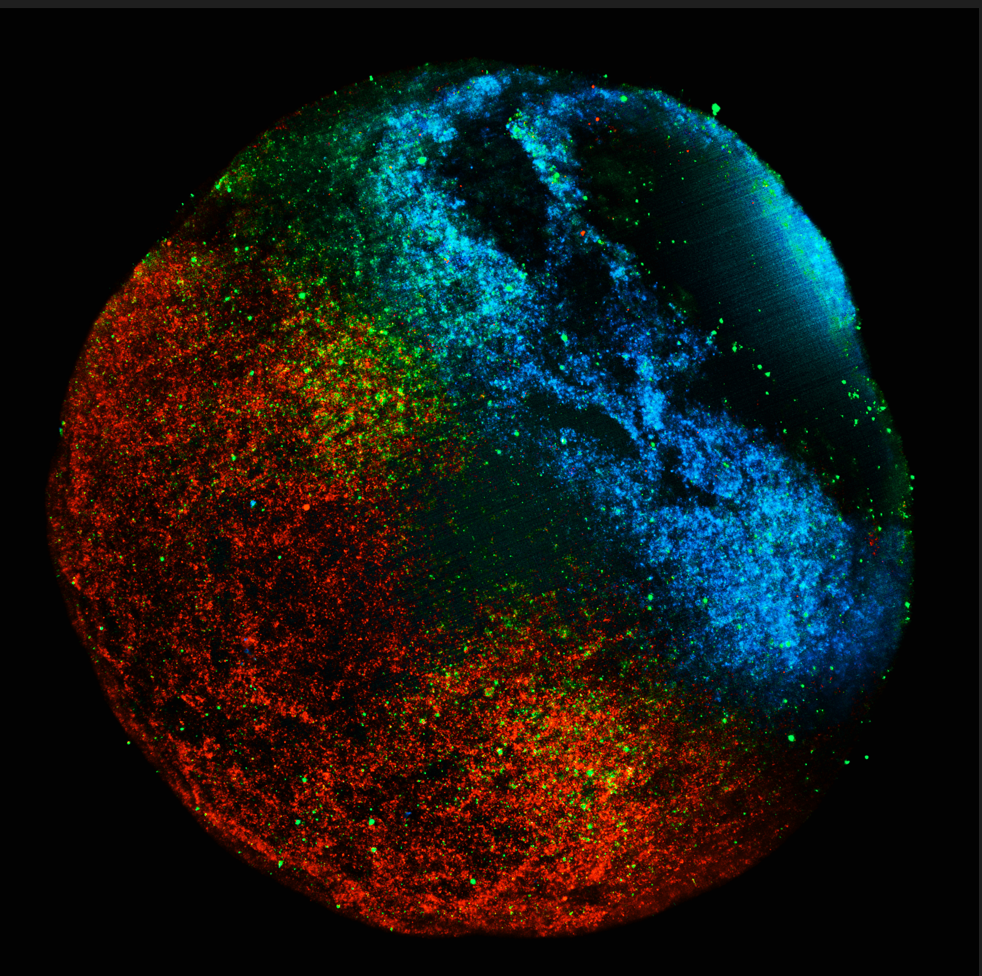
How did you acquire the images used in the documentary/exhibition? So, one of the videos that was featured, and perhaps the video I’m most proud of, is the timelapse showing the difference in the invasion capacity between two lines of melanoma cells, one is labelled in magenta and the other in cyan. The cells were dyed separately with two types of cell tracker, one red and one green but I used Fiji to change the colours into magenta and cyan, which are more colour-blind friendly. The two cell populations are then mixed in a 1 to 1 ratio and seeded to confluence in a 6-well plate coated with fibronectin. A wound was created by scratching with a pipette tip for the cells to migrate into. This gap was imaged with a Nikon TE2000 microscope equipped with a PlanFluor 10x/0.30 objective every 15min for 20h in both brightfield (not shown) and fluorescence at the excitation wavelength of 488 and 568nm in a 5% CO2 humidified chamber.
What do you think is the most exciting innovation in microscopy? I think this is definitely light sheet microscopy. The kind of spatiotemporal dynamics that you can get out of this is incredible. I felt in love with it when I saw those amazing videos of T-cells making contacts and releasing granzyme to kill cancer cells from the lab of Gillian Griffith in Cambridge. I really do hope I will be able to learn it in the future.
Alex McDougall
Research: My research focusses on how embryo shape emerges during the earliest part of development when zygotic genome activation is either absent or minimal. Most biologists would be been able to identify early embryos of different species (eg. 16/32 cell stages), but if asked how these shapes emerge then they would find that a more difficult question to answer. That is because the emergence of embryo shape arises from the interplay (for example and not an exhaustive list) between cell biological mechanisms and biomechanical properties that depend largely on proteins/RNAs/organelles/biomechanical properties that control cell position (through cell division orientation), cell shape (cortical tension and cell adhesion) and ultimately embryo shape. To study this question we have been exploiting the animal that displays the most invariant cleavage pattern of all animals – the ascidian. Specifically we use the embryo of Phallusia mammillata – an emergent model organism that was chosen in the 1980s by Christian Sardet at the Villefranche Marine Station IMEV and further developed by myself at IMEV starting in the 1990s and then several other laboratories for research – Phallusia is useful for microsopy studies because of its transparent eggs/embryos.

How did you become involved with the project? We were contacted by Terence and Renaud who found us on Twitter.
What do you think about Cell Worlds? I love the project – bravo Terence and Renaud
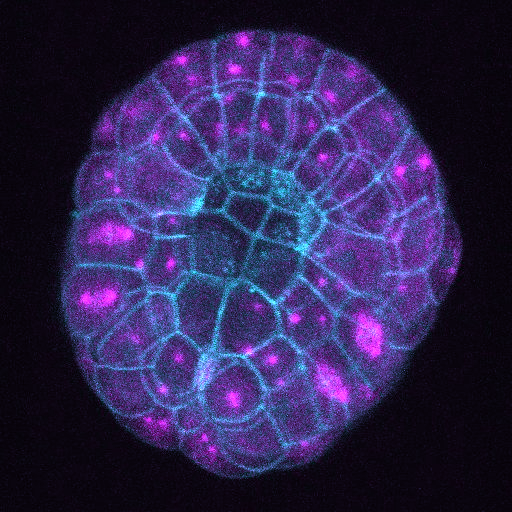
How did you acquire the images used in the documentary/exhibition? Via confocal 4D microscopy (Leica SP8 with Hybrid detector). Images were generally acquired every 2 min. with up to 50 z planes and about 1.5 µm step size.
Since we submitted many movies the imaging parameters above are generic and could be slightly different from movie to movie – all however are confocal 4D.
Noemie Paillon
Research: PD-1 blockade is a promising immunotherapy used against several cancers. PD-1 binding to its ligand PD-L1, often expressed on tumor cells, dampens T cells activation and prevents tumor clearance. Further understanding the mechanisms underlying PD-1-induced T cell inhibition could allow for enhanced therapeutic success. My research project therefore aims at deciphering how PD-1 alters the formation of an immune synapse between T cells and cancer cells.

How did you become involved with the project? I posted a “dancing T cell” on Twitter that caught the attention of the Cell Worlds team. I was immediately enthusiastic to join the project, since I always felt like the beauty of microscopy could amaze the public beyond scientists. Hence, I am grateful to Renaud and Terence for giving me the opportunity to make my cells travel around the world.
What do you think about Cell Worlds? The Cell Worlds team managed to assemble diverse raw microscopy data into a unique project, successfully immersing us in the amazing world within our body. Not only does it bring together various aspects of biology, the project also creates a bridge between microscopists and citizens.
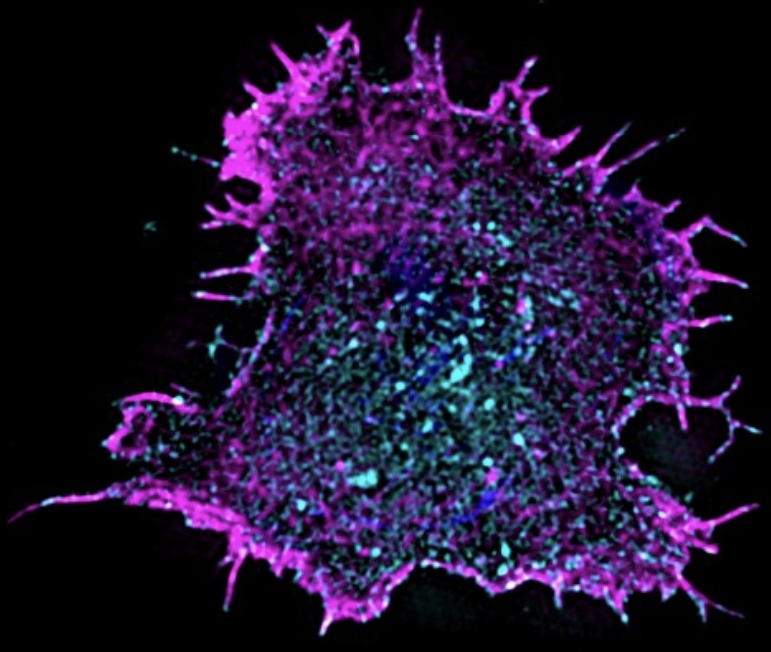
How did you acquire the images used in the documentary/exhibition? My “dancing T cell” was imaged in 4D using a lattice light-sheet microscope, enabling us to observe it sensing its environment with actin protrusions. A second set of images appearing in the documentary shows the actin cytoskeleton (magenta) and PD-1 (cyan) of spreading T cells that were acquired on a 3D-SIM microscope.
What do you think is the most exciting innovation in microscopy? Lattice light-sheet microscopy is an amazing tool to follow cells in a precise 3D reconstruction and high speed. It opens opportunities to better grasp how cells communicate and function in a dynamic manner.
Martyna Lukoseviciute
Research: The images that I shared for the ‘Cell Worlds’ were acquired during my PhD work. I was working on zebrafish embryo development, specifically, on chromatin dynamics and cellular heterogeneity underlying multipotent neural crest development. For my current postdoctoral adventure, I have migrated from developmental biology to regeneration field. I am trying to shed light on how immune system contributes to neural stem cell activation and differentiation after central nervous system injuries.

How did you become involved with the project? I started employing microscopy methods during my research and quickly fell in love with imaging. I think this was a very natural development for me because I always had an artistic side, hence I found that microscopy could help me to marry two of my favourite worlds – art and science. Additionally, after sharing some of my images with family and friends I realised that microscopic images appeal not only to scientists, but also to a wider public, thus such images can be used as tools for public engagement. Therefore, I was very excited when Renaud approached me on Twitter after seeing one of my tweets showing an image of a developing zebrafish embryo and invited me to participate in the ‘Cell Worlds’ project. The vision of this Art and Science project aiming to raise public awareness and curiosity of the microscopic world directly aligned with my own interests. I thought it was a great initiative and so I was happy to contribute with my images.
What do you think about Cell Worlds? The project’s aim was to unlock the doors to the microscopic world to the general public via art and science communication. I think that it was a great and successful idea to show the images and videos in two different ways – the exhibition and the documentary. The exhibition allowed the viewers to fully immerse themselves into this unknown world, to become a part of it, to see it from different angles and in a great resolution. While the documentary allowed the viewers to not only experience this world, but also to understand it better. I was very impressed how ‘Cell Worlds’ team managed to combine images and videos that they have received from different researchers into a coherent, engaging, and educational story. I heard that the documentary was very appealing to children, and I think this is the most genuine success of the project – to inspire the new generation for science, to ignite their curiosity and explorative side, and to show that science can be very colourful and exciting.
How did you acquire the images used in the documentary/exhibition? I used Zeiss LSM 780 inverted confocal microscope equipped with Plan-APO Chromat 10x/0.45 NA WD=2.0 mm or Zeiss Lighsheet Z.1 microscope equipped with 10x/0.5 NA water dipping detection objective.
What do you think is the most exciting innovation in microscopy? Even though I have not used it myself yet, I think that a new lattice light-sheet microscopy technique is very exciting and enables unlocking previously inaccessible knowledge, i.e., imaging very dynamic and fast processes at high-resolution in live samples with little phototoxicity.
Edouard Hannezo and Tsuyoshi Hirashima
Research: Edouard – Our group is interested in understanding how cells “know” how to behave at the right place and at the right time during embryo development or tissue homeostasis. We use methods from theoretical physics (e.g. solid and fluid mechanics, statistical physics or soft matter) to understand how these choices occur in a robust manner.

Tsuyoshi – Our group is interested in how collective cell behavior emerges and links with biological functions in development and reproduction. We focus on cellular mechano-chemical coupling regulations in mammalian tissues and explore design principles of multicellular dynamical systems using live cell imaging, modeling, and simulations.

How did you become involved with the project? We posted some movies from our work on Twitter when we published our joint work (Boocock*, Hino* et al., Nat Phys, 2021). Renaud and Terence found those and contacted us to know whether they could use some of them. This got us of course really excited, especially when we heard about the project
What do you think about Cell Worlds? This stunning project has brought people on fantastic journeys with great excitement. It’s really cool for us to be side-by-side with so many cool works and scientists in this exhibit. Congratulations and kudos to the team for combining so well art and science, and looking forward to your exciting future activities.
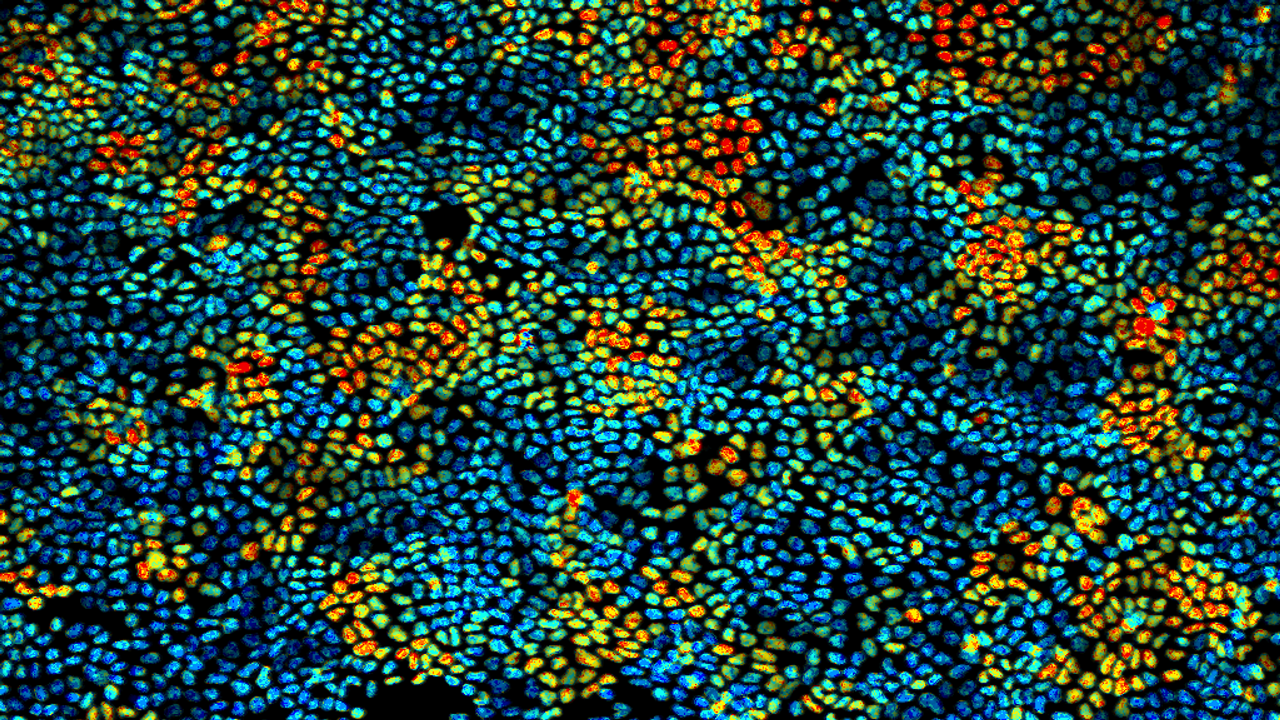
How did you acquire the images used in the documentary/exhibition? Our images show ERK/MAP kinase activity in nuclei of MDCK cells under the confluent condition. The ERK activity was visualized by FRET imaging for a genetically-encoded biosensor using either a widefield or a confocal microscope system.
What do you think is the most exciting innovation in microscopy? Digital image acquisition using image sensors and the A/D converter is one of the innovations in the past, which enabled data processing to help researchers recognize what and how cells of interest are. A possible future of microscopy is a way to provide more tangible experiences for the users with the power of Metaverse technologies. This will allow us to immerse into beautiful microscopic Cell Worlds, which will change the current style of microscope systems.
Paul Guichard and Virginie Hamel
Research: Our laboratory focuses on deciphering the structural mechanisms governing centriole assembly and architecture. To this end, we are combining cell biology methods, in vitro reconstitution assays, cryo-microscopy/cryo-tomography and super-resolution microscopy including expansion microscopy to gain decisive insights into these mechanisms.

How did you become involved with the project? We have been contacted by Renaud and Terence, the co-founders of Cell Worlds, asking whether we would be happy to contribute to their exciting project and provide a couple of beautiful images. We have been very impressed by their project and dedication to it and are extremely grateful for being part of this adventure, also our contribution was minimal compared to other labs.
What do you think about Cell Worlds? The project was extremely well thought, designed, and executed. It is incredibly beautiful and useful to engage with the public.
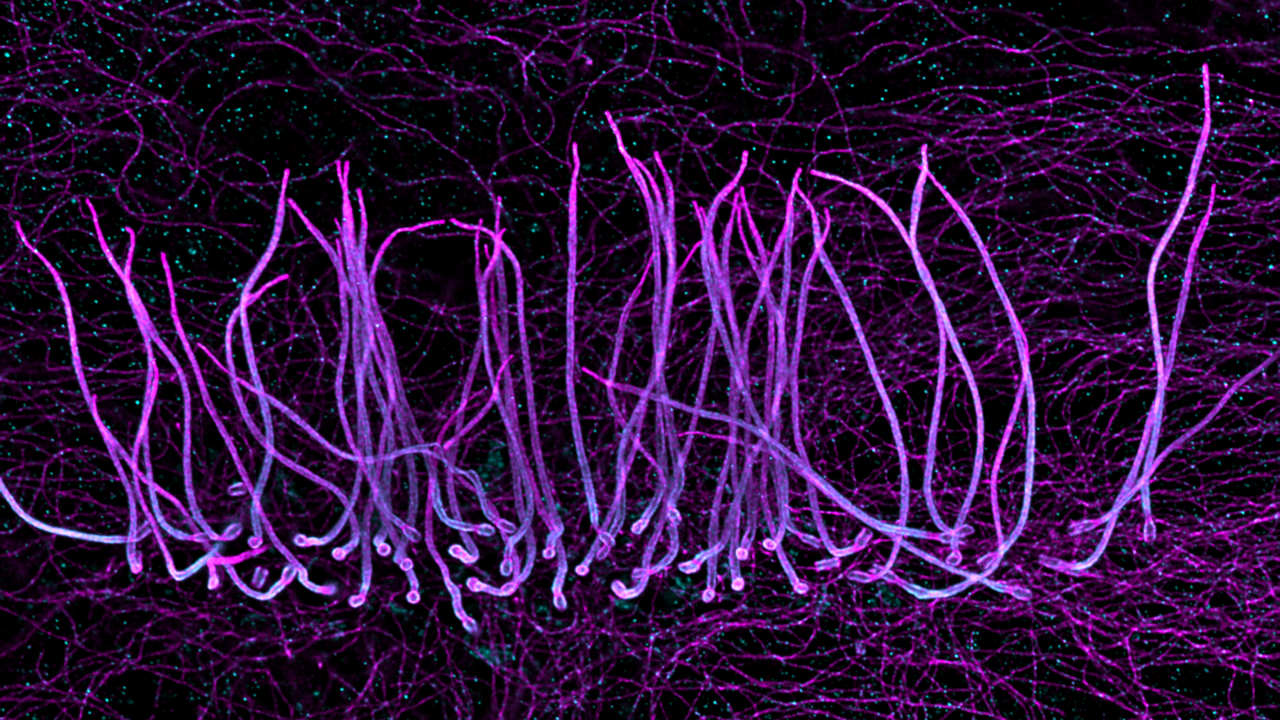
How did you acquire the images used in the documentary/exhibition? The images provided by our laboratory were taken from biological samples that were expanded 4-fold to acquire super-resolution images using a conventional Leica SP8 confocal. One image represents a human cell stained for microtubules and highlighting a primary cilium, that can be seen as the cell antenna. The other image is taken from ependymal ciliated cells and illustrate the organization of motile cilia.
What do you think is the most exciting innovation in microscopy? One key innovation in microscopy is the invention of the expansion microscopy technique by Ed Boyden in 2015 that gave rise now to many optimized or dedicated protocols for specific applications. The principle of this technique relies on the physical expansion of the biological specimen and allows super-resolution imaging to all laboratories.
The lab of Jean-Leon Maitre
Research: We are interested in the shaping of the blastocyst, the structure formed by mammalian embryos during the first few days of development. To better understand blastocyst morphogenesis, we use microscopy to see the shape changes, biophysics to identify changes in material properties associated with these shape changes, and genetics to disrupt all of these.

How did you become involved with the project? Terence and Renaud contacted Jean-Léon, who immediately saw this as a unique possibility to share our beautiful images to the public. Terence and Renaud came to visit the lab and discussed with all of us, so they could pick directly their favourite images and movies.
What do you think about Cell Worlds? It feels obvious to us that microscopy images of living organisms are stunningly beautiful but it took the insights and craft of Cell Worlds to make the images accessible to the public. The characters of Terence and Renaud were certainly a prime determinant in the ability of Cell Worlds to collect so many beautiful images from labs all over the planet. Twitter probably helped as well!
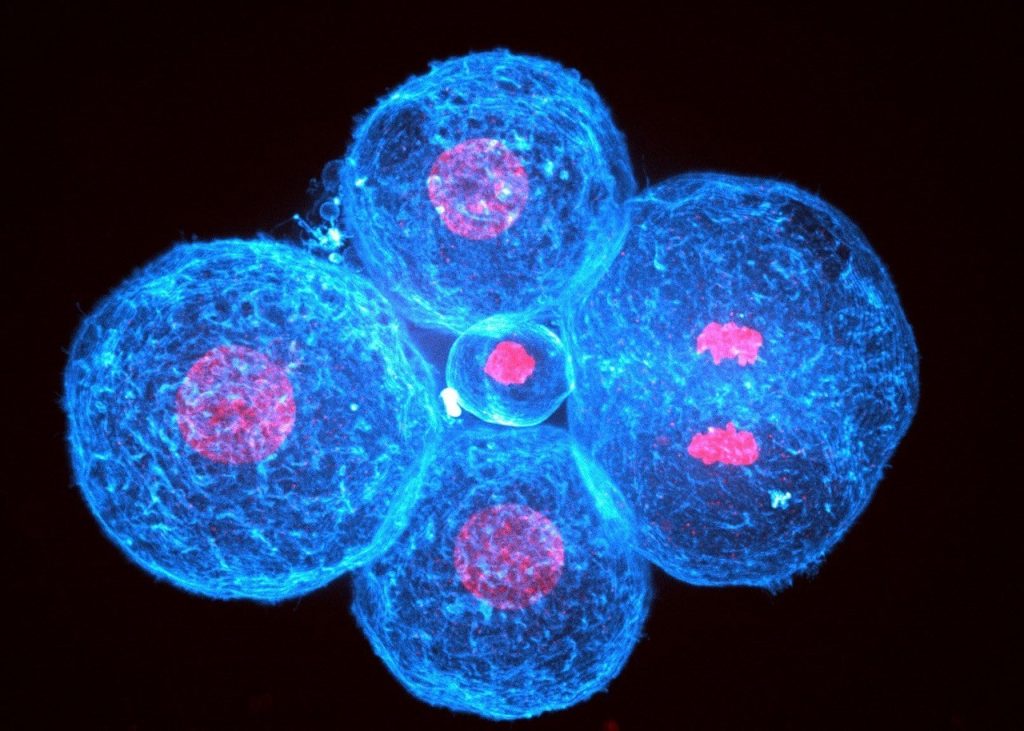
How did you acquire the images used in the documentary/exhibition? The images we provided come either from spinning disc or light sheet confocal microscopy. Because mammalian embryos develop slowly and internally, they are very sensitive to photodamage and we have to be very gentle when we image them.
What do you think is the most exciting innovation in microscopy? JLM: Lattice light sheet microscopy certainly seems appealing but I feel that recently the biggest technological jumps have come from computational treatments of images. The image analysis community is extremely friendly and open, which is most likely a major reason for this success.
Thanks to Anh, Alex, Noemie, Martyna, Tsuyoshi & Edouard, Virginie & Paul and the lab of Jean-Leon Maitre for a giving us some insight into their work and the story behind the images featured in Cell Worlds. If you haven’t already seen them, check out the Cell Worlds documentary and the interview with Cell Worlds founders, Renaud Pourpre and Terence Saulnier on our sister site, the Node.


 (No Ratings Yet)
(No Ratings Yet)