Microscopy preprints – applications in cell biology
Posted by FocalPlane, on 12 August 2022
Here is a curated selection of preprints published recently. In this post, we focus specifically on preprints using microscopy tools in cell biology.
Cryo-electron tomography reveals enrichment and identifies microtubule lumenal particles in neuronal differentiation
Saikat Chakraborty, Antonio Martinez-Sanchez, Florian Beck, Mauricio Toro-Nahuelpan, In-Young Hwang, Kyung-Min Noh, Wolfgang Baumeister, Julia Mahamid

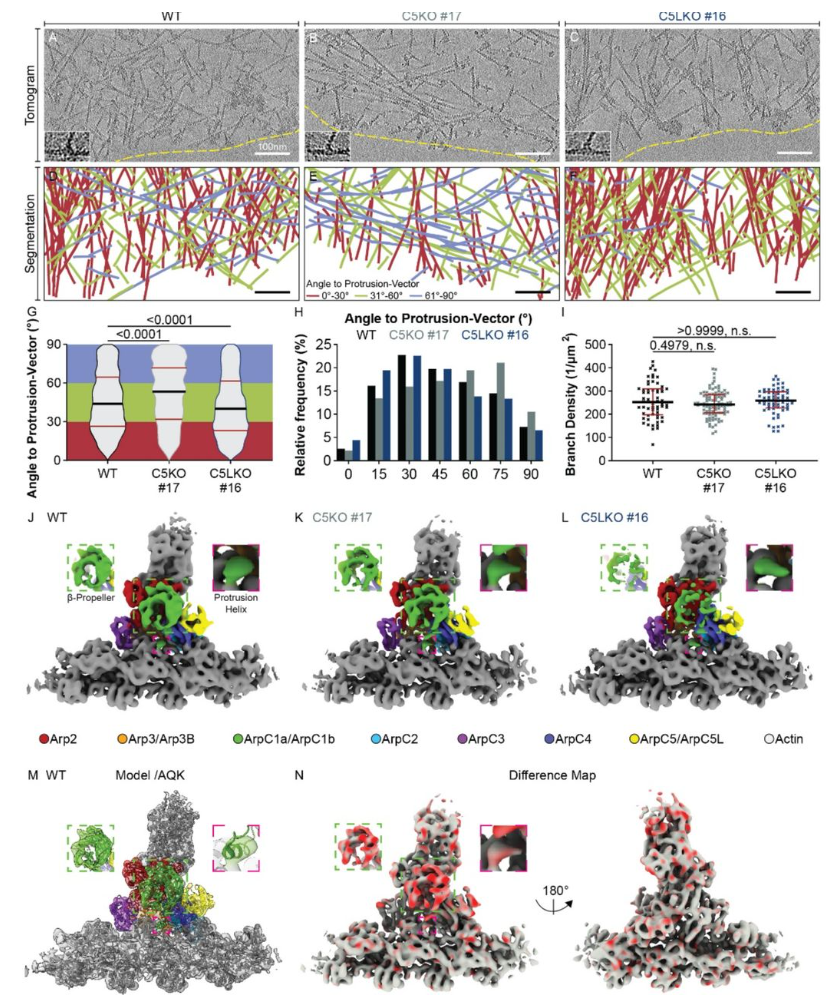
ArpC5 isoforms regulate Arp2/3 complex-dependent protrusion through differential Ena/VASP positioning
Florian Fäßler, Manjunath G Javoor, Julia Datler, Hermann Döring, Florian W Hofer, Georgi Dimchev, Victor-Valentin Hodirnau, Klemens Rottner, Florian KM Schur
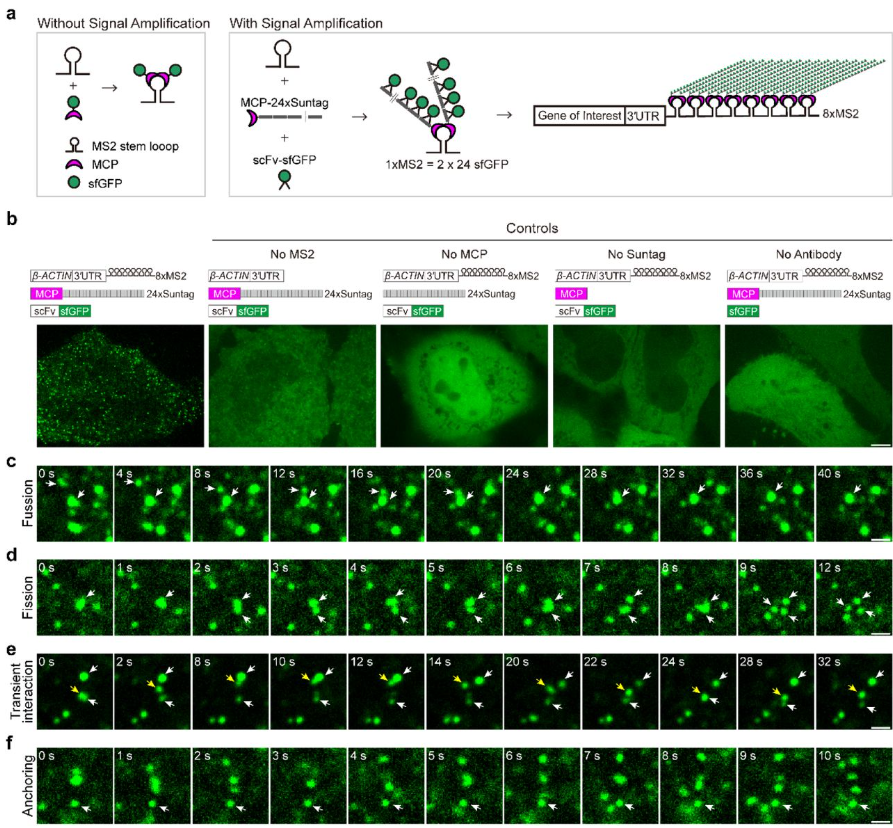
Enhanced Single RNA Imaging Reveals Dynamic Gene Expression in Live Animals
Yucen Hu, Jingxiu Xu, Erqing Gao, Xueyuan Fan, Jieli Wei, Suhong Xu, Weirui Ma
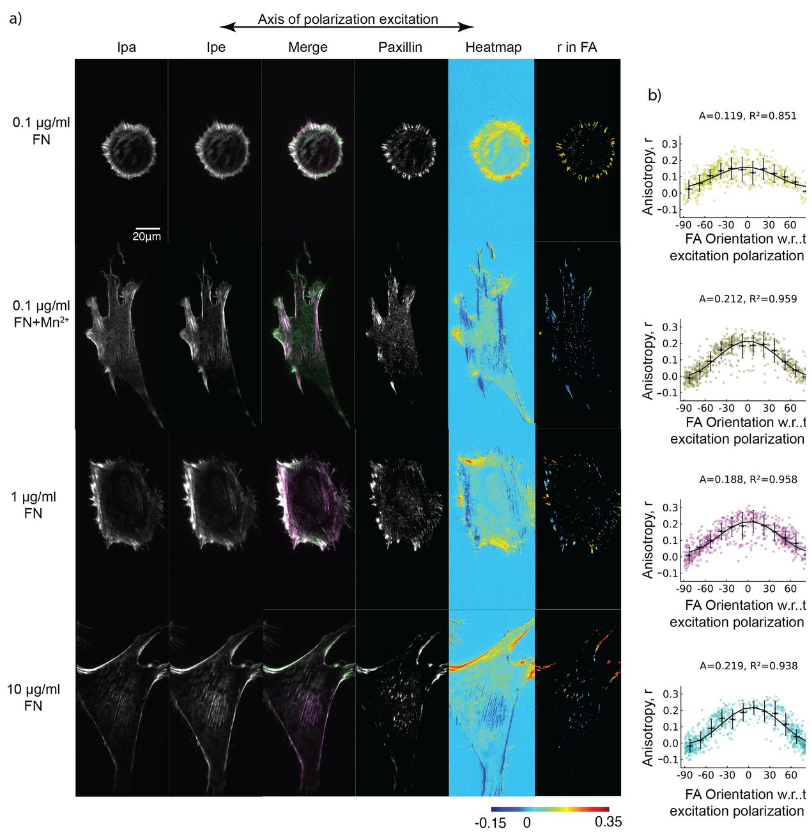
Extracellular matrix sensing via modulation of orientational order of integrins and F-actin in focal adhesions
Valeriia Grudtsyna, Swathi Packirisamy, Tamara Bidone, Vinay Swaminathan
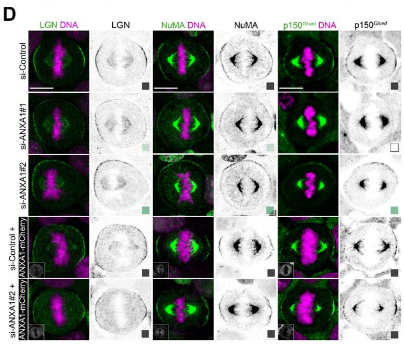
Annexin A1 is a polarity cue that directs planar mitotic spindle orientation during mammalian epithelial morphogenesis
Maria Fankhaenel, Farahnaz Sadat Golestan Hashemi, Larissa Mourao, Emily Lucas, Manal Mosa Hosawi, Paul Skipp, Xavier Morin, Colinda L.G.J. Scheele, Salah Elias
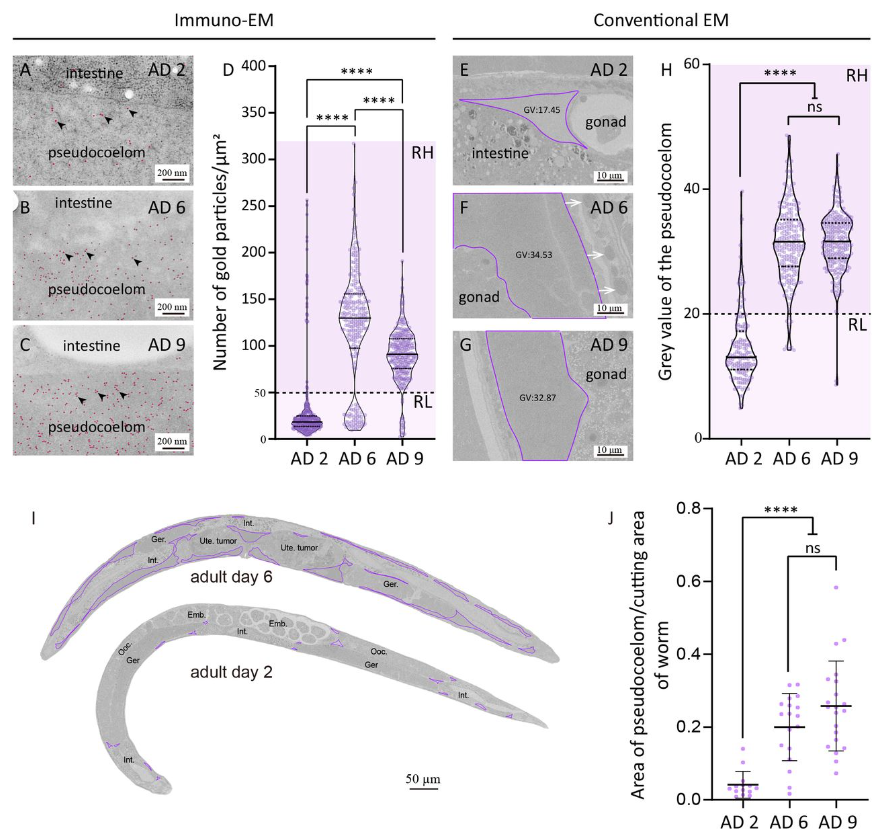
Old Age Drives Fusion and Expansion of Vitellogenin Vesicles in the Intestine of Caenorhabditis elegans
Chao Zhai, Nan Zhang, Xi-Xia Li, Xi Chen, Fei Sun, Meng-Qiu Dong
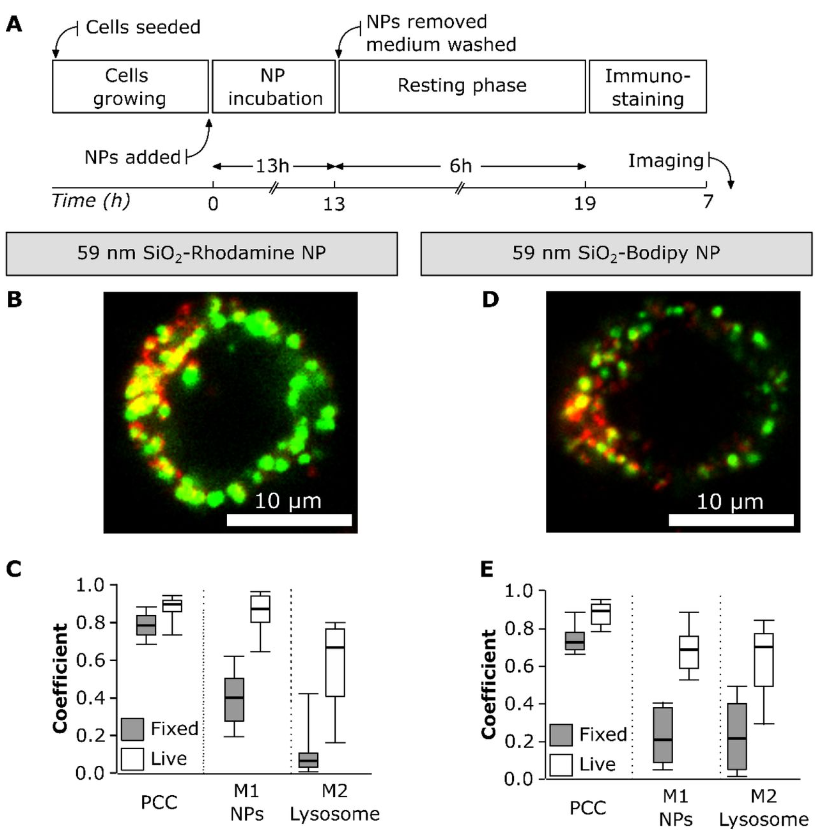
Pitfalls in methods to study colocalization of nanoparticles in mouse macrophage lysosomes
Aura Maria Moreno-Echeverri, Eva Susnik, Dimitri Vanhecke, Patricia Taladriz-Blanco, Sandor Balog, Alke Petri-Fink, Barbara Rothen-Rutishauser
Experimental determination and mathematical modeling of standard shapes of forming autophagosomes
Yuji Sakai, Satoru Takahashi, Ikuko Koyama-Honda, Chieko Saito, Noboru Mizushima

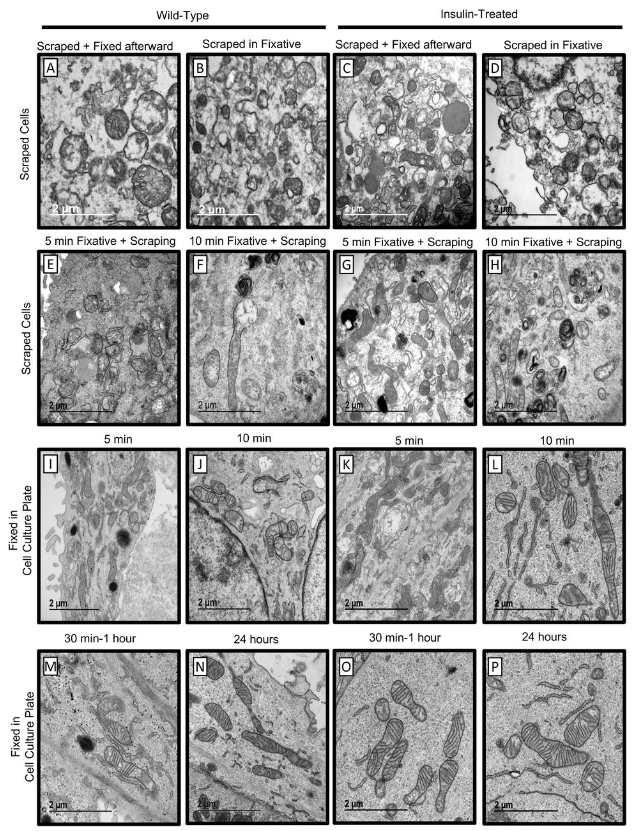
A comprehensive approach to artifact-free sample preparation and the assessment of mitochondrial morphology in tissue and cultured cells
Antentor Hinton Jr., Prasanna Katti, Trace A. Christensen, Margaret Mungai, Jianqiang Shao, Liang Zhang, Sergey Trushin, Ahmad Alghanem, Adam Jaspersen, Rachel E. Geroux, Kit Neikirk, Michelle Biete, Edgar Garza Lopez, Zer Vue, Heather K. Beasley, Andrea G. Marshall, Jessica Ponce, Christopher K. E. Bleck, Innes Hicsasmaz, Sandra A. Murray, Ranthony A.C. Edmonds, Andres Dajles, Young Do Koo, Serif Bacevac, Jeffrey L. Salisbury, Renata O. Pereira, Brian Glancy, Eugenia Trushina, E. Dale Abel
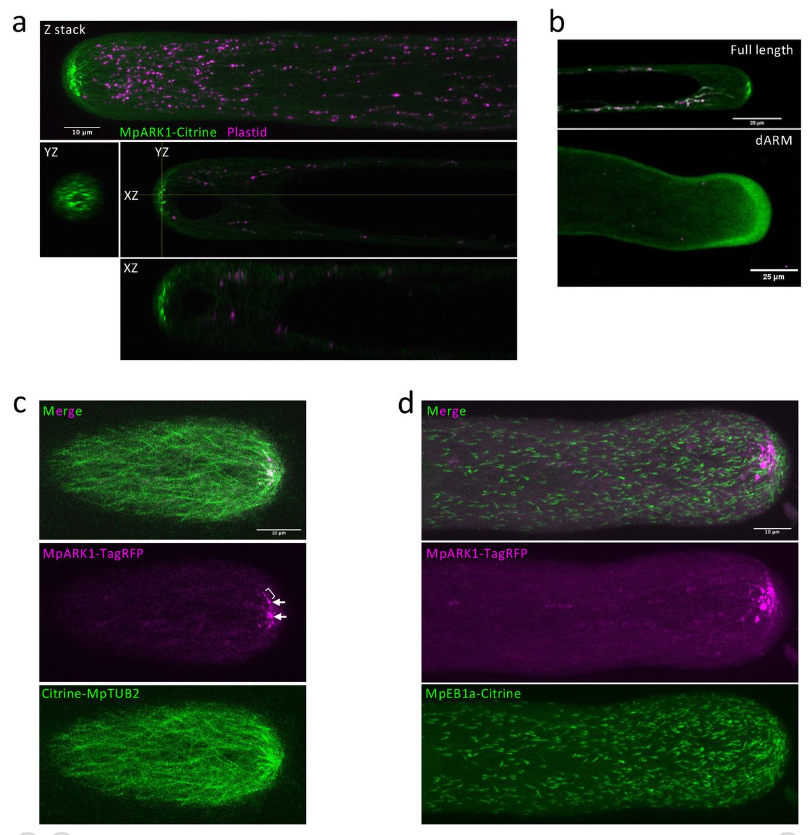
Plant specific armadillo repeat kinesin directs organelle transport and microtubule convergence to promote tip growth
Asaka Kanda, Kento Otani, Taku Takahashi, Hiroyasu Motose
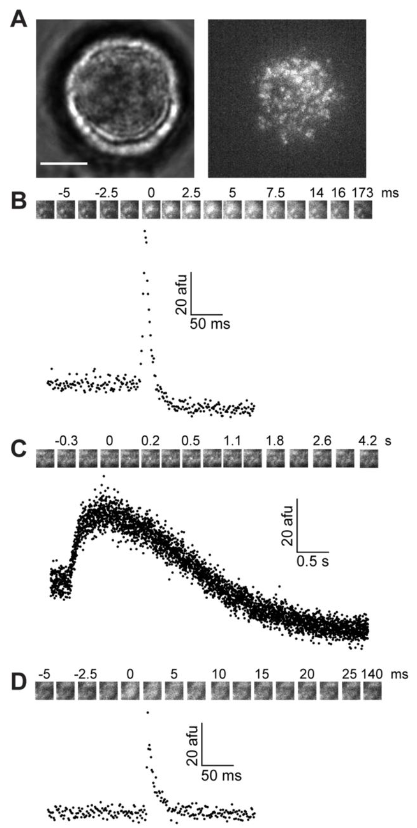
High-Speed Imaging Reveals the Bimodal Nature of Dense Core Vesicle Exocytosis
Pengcheng Zhang, David Rumschitzki, Robert H. Edwards
Cellular Uptake of His-Rich Peptide Coacervates Occurs by a Macropinocytosis-Like Mechanism
Anastasia Shebanova, Quentin Perrin, Sushanth Gudlur, Yue Sun, Zhi Wei Lim, Ruoxuan Sun, Sierin Lim, Alexander Ludwig, Ali Miserez
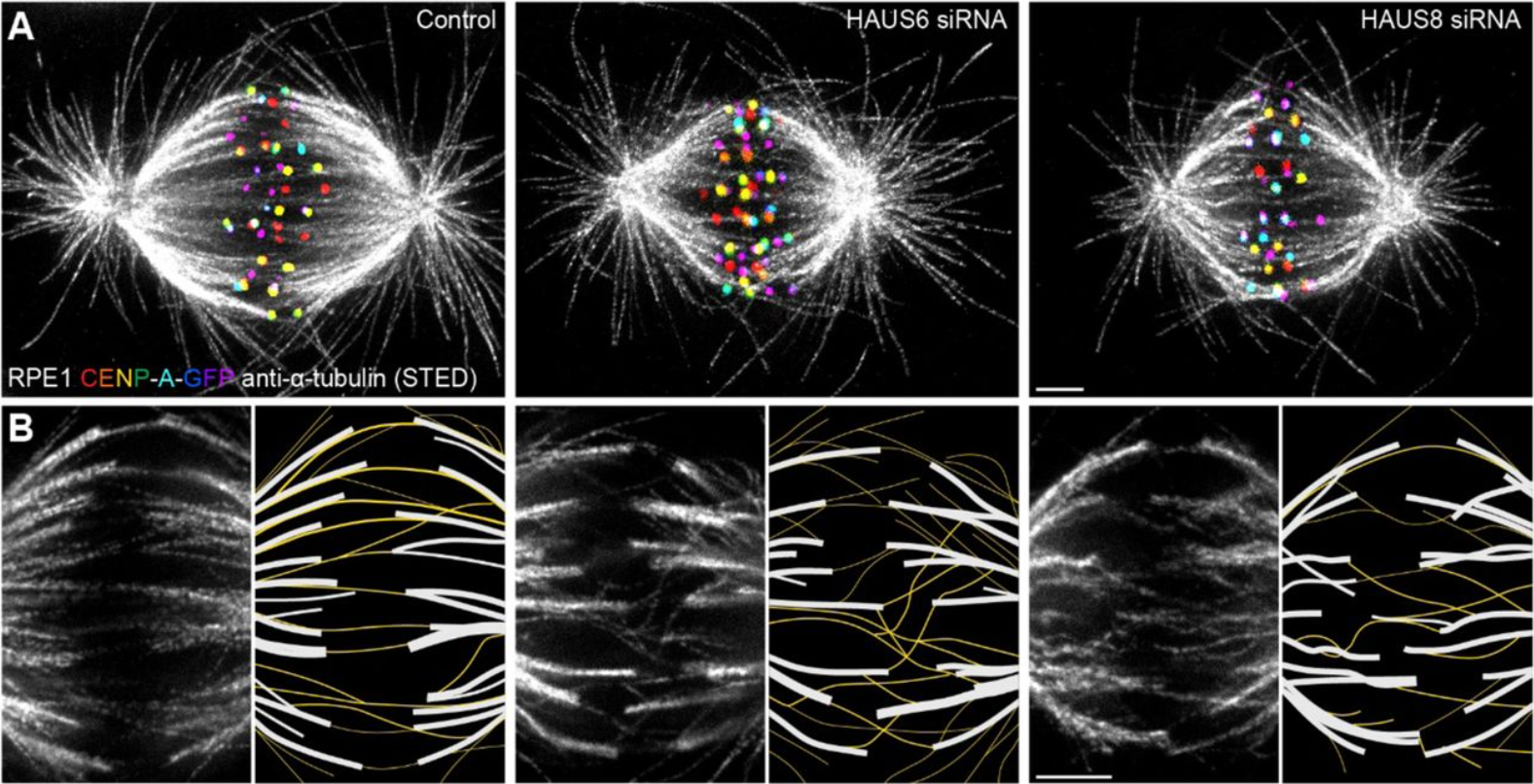
Augmin prevents merotelic attachments by promoting proper arrangement of bridging and kinetochore fibers
Valentina Štimac, Isabella Koprivec, Martina Manenica, Juraj Simunić, Iva M. Tolić
True-to-scale DNA-density maps correlate with major accessibility differences between active and inactive chromatin
Márton Gelléri, Shih-Ya Chen, Aleksander Szczurek, Barbara Hübner, Michael Sterr, Jan Neumann, Ole Kröger, Filip Sadlo, Jorg Imhoff, Yolanda Markaki, Michael J. Hendzel, Marion Cremer, Thomas Cremer, Hilmar Strickfaden, Christoph Cremer
Biophysical ordering transitions underlie genome 3D re-organization during cricket spermiogenesis
Guillermo A. Orsi, Maxime M.C. Tortora, Béatrice Horard, Dominique Baas, Jean-Philippe Kleman, Jonas Bucevičius, Gražvydas Lukinavičius, Daniel Jost, Benjamin Loppin
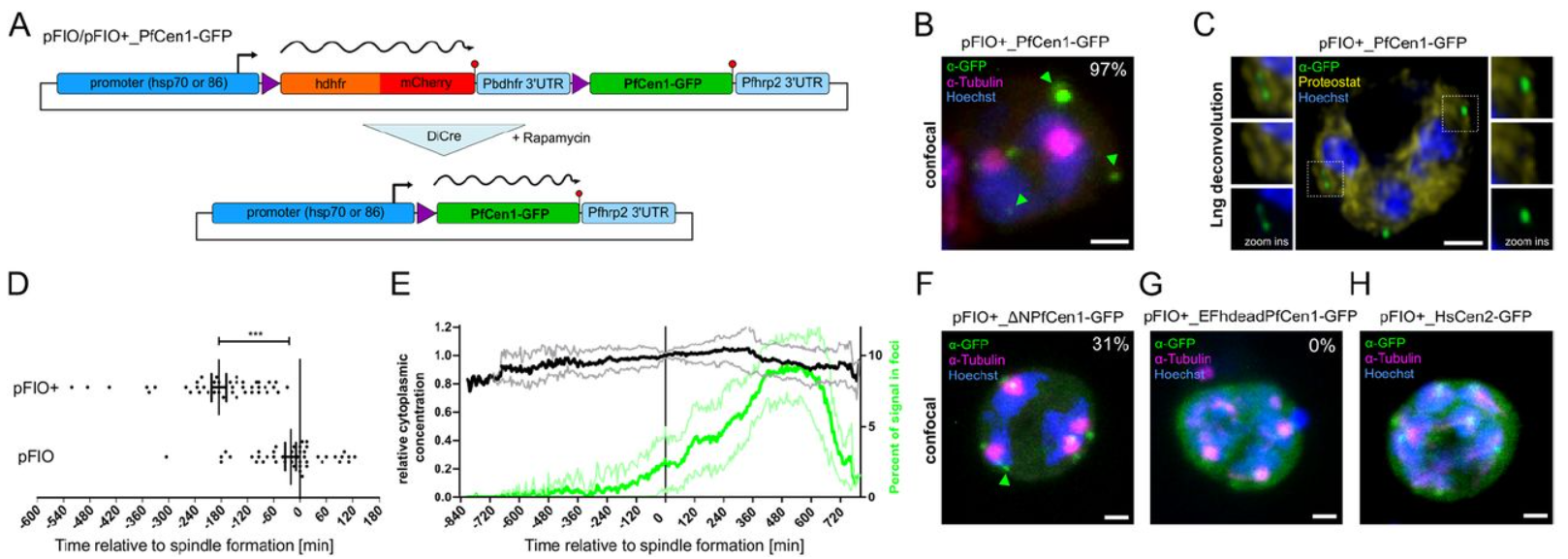
Ca2+-inducible phase separation of centrins in proliferating malaria parasites
Yannik Voß, Severina Klaus, Markus Ganter, Julien Guizetti
Helical metaphase chromatid coiling is conserved
Ivona Kubalová, Amanda Souza Câmara, Petr Cápal, Tomáš Beseda, Jean-Marie Rouillard, Gina Marie Krause, Helena Toegelová, Axel Himmelbach, Nils Stein, Andreas Houben, Jaroslav Doležel, Martin Mascher, Hana Šimková, Veit Schubert
Architecture and dynamics of a novel desmosome-endoplasmic reticulum organelle
Navaneetha Krishnan Bharathan, William Giang, Jesse S. Aaron, Satya Khuon, Teng-Leong Chew, Stephan Preibisch, Eric T. Trautman, Larissa Heinrich, John Bogovic, Davis Bennett, David Ackerman, Woohyun Park, Alyson Petruncio, Aubrey V. Weigel, Stephan Saalfeld, COSEM Project Team, A. Wayne Vogl, Sara N. Stahley, Andrew P. Kowalczyk
Long-term mitotic DNA damage promotes chromokinesin-mediated missegregation of polar chromosomes in cancer cells
Marco Novais-Cruz, António Pombinho, Mafalda Sousa, André F. Maia, Helder Maiato, Cristina Ferrás
Both ANT and ATPase are essential for mitochondrial permeability transition but not depolarization
M.A. Neginskaya, S.E. Morris, E.V. Pavlov

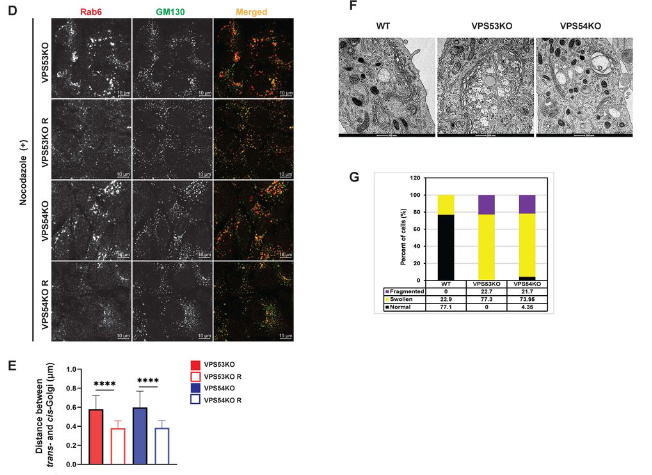
GARP complex controls Golgi physiology by stabilizing COPI machinery and Golgi v-SNAREs
Amrita Khakurel, Tetyana Kudlyk, Irina Pokrovskaya, Zinia D’Souza, Vladimir V. Lupashin
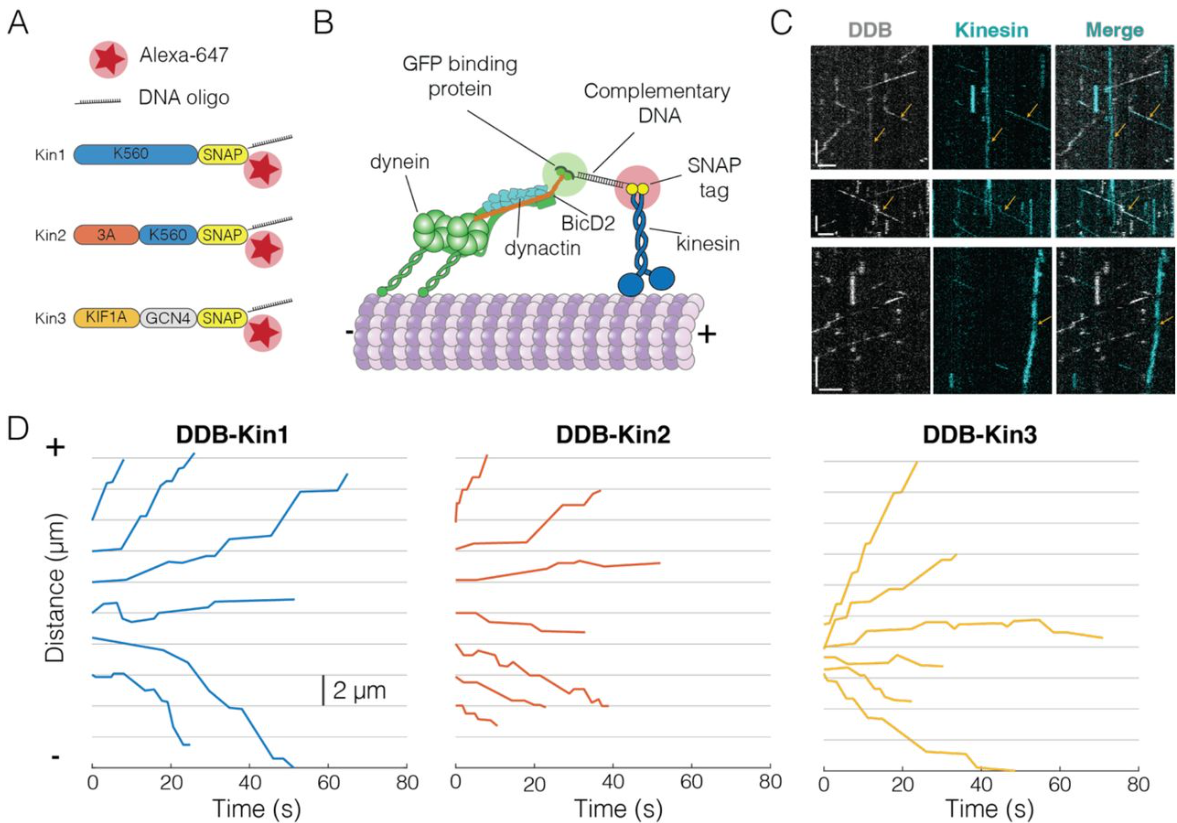
Kinesin-1, -2 and -3 motors use family-specific mechanochemical strategies to effectively compete with dynein during bidirectional transport
Allison M. Gicking, Tzu-Chen Ma, Qingzhou Feng, Rui Jiang, Somayesadat Badieyan, Michael A. Cianfrocco, William O. Hancock
Development of a fluorescence reporter system to quantify transcriptional activity of endogenous p53 in living cells
Tatsuki Tsuruoka, Emiri Nakayama, Takuya Endo, Shingo Harashima, Rui Kamada, Kazuyasu Sakaguchi, Toshiaki Imagawa


 (No Ratings Yet)
(No Ratings Yet)