Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 26 August 2022
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools only.
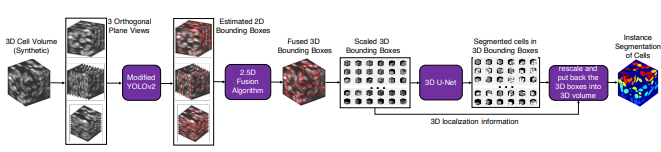
YOLO2U-Net: Detection-Guided 3D Instance Segmentation for Microscopy
Amirkoushyar Ziabari, Derek C. Ros, Abbas Shirinifard, David Solecki
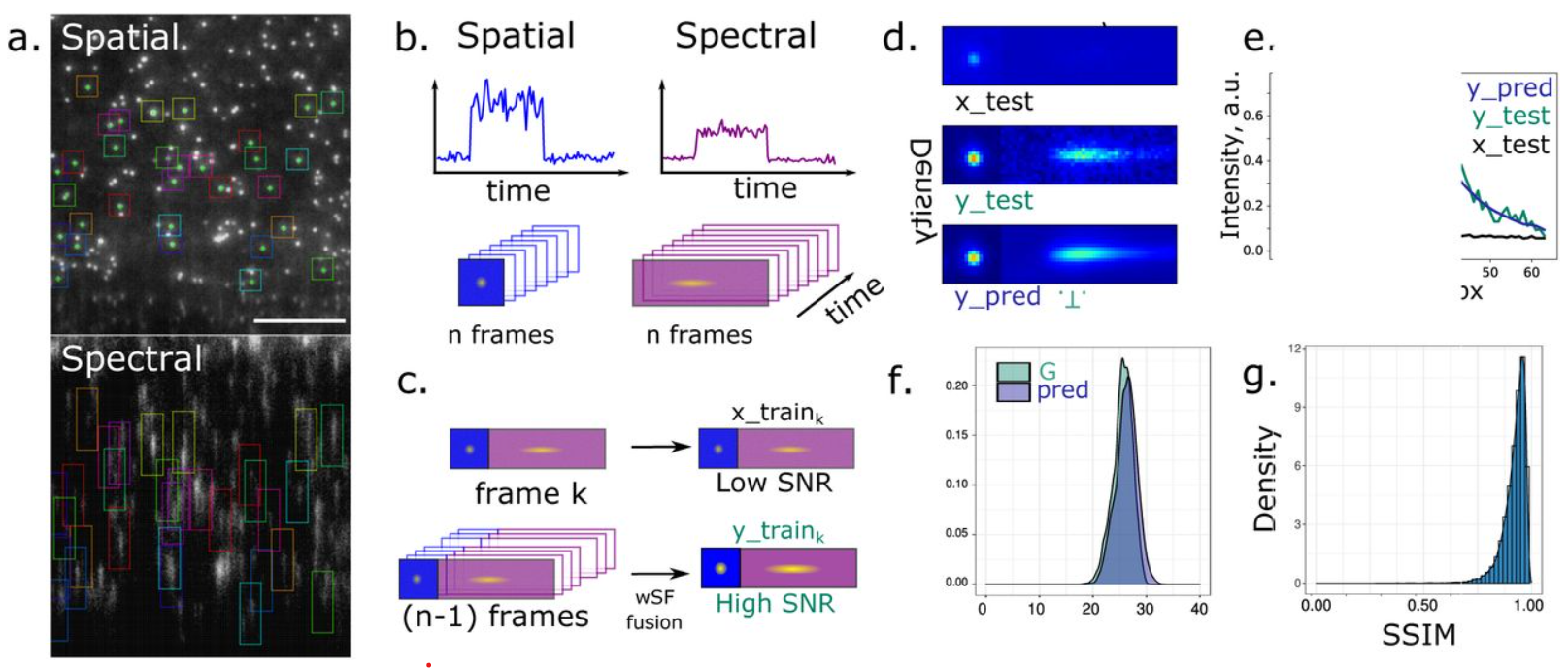
Advancing Spectrally-Resolved Single Molecule Localization Microscopy using Deep Learning
Hanna Manko, Yves Mély, Julien Godet
u-track 3D: measuring and interrogating dense particle dynamics in three dimensions
Philippe Roudot, Wesley R. Legant, Qiongjing Zou, Kevin M. Dean, Tadamoto Isogai, Erik S. Welf, Ana F. David, Daniel W. Gerlich, Reto Fiolka, Eric Betzig, Gaudenz Danuser
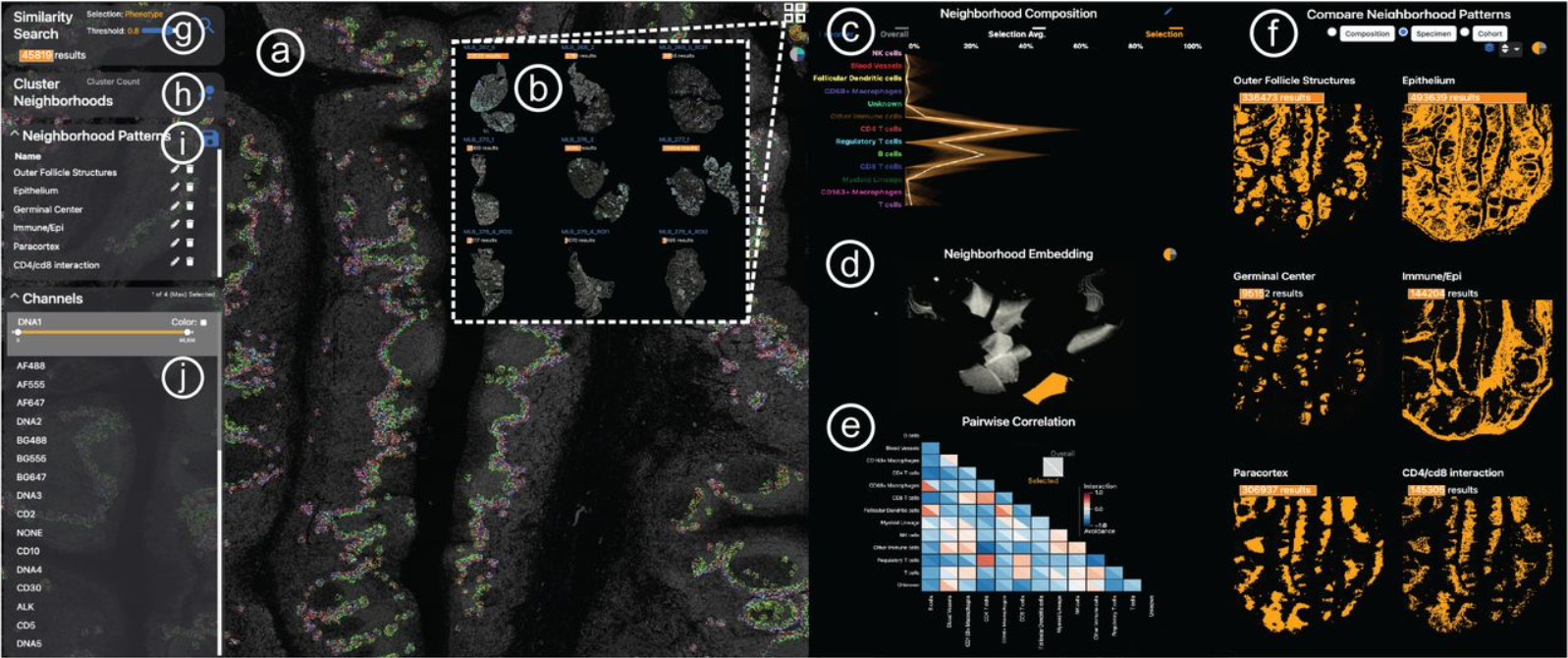
Visinity: Visual Spatial Neighborhood Analysis for Multiplexed Tissue Imaging Data
Simon Warchol, Robert Krueger, Ajit Johnson Nirmal, Giorgio Gaglia, Jared Jessup, Cecily C. Ritch, John Hoffer, Jeremy Muhlich, Megan L. Burger, Tyler Jacks, Sandro Santagata, Peter K. Sorger, Hanspeter Pfister
Precise targeting for 3D cryo-correlative light and electron microscopy volume imaging of tissues using a FinderTOP
Marit de Beer, Deniz Daviran, Rona Roverts, Luco Rutten, Elena Macías Sánchez, Jurriaan R. Metz, Nico Sommerdijk, Anat Akiva
Cell segmentation from telecentric bright-field transmitted light microscopy images using a Residual Attention U-Net: a case study on HeLa line
Ali Ghaznavi, Renata Rychtarikova, Mohammadmehdi Saberioon, Dalibor Stys
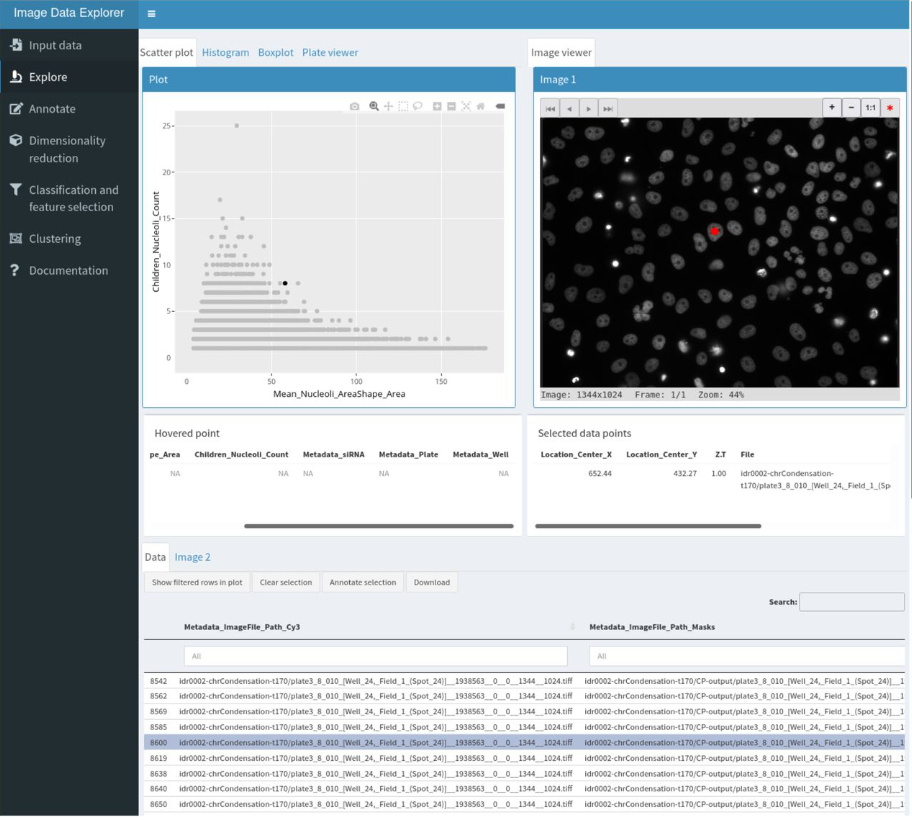
The Image Data Explorer: interactive exploration of image-derived data
Coralie Muller, Beatriz Serrano-Solano, Yi Sun, Christian Tischer, Jean-Karim Hériché
Deep neural network automated segmentation of cellular structures in volume electron microscopy
Benjamin Gallusser, Giorgio Maltese, Giuseppe Di Caprio, Tegy John Vadakkan, Anwesha Sanyal, Elliott Somerville, Mihir Sahasrabudhe, Justin O’Connor, Martin Weigert, Tom Kirchhausen
MEPSi: A tool for simulating tomograms of membrane-embedded proteins
Borja Rodríguez de Francisco, Armel Bezault, Xiao-Ping Xu, Dorit Hanein, Niels Volkmann
3D Centroidnet: Nuclei Centroid Detection with Vector Flow Voting
Liming Wu, Alain Chen, Paul Salama, Kenneth W. Dunn, Edward J. Delp
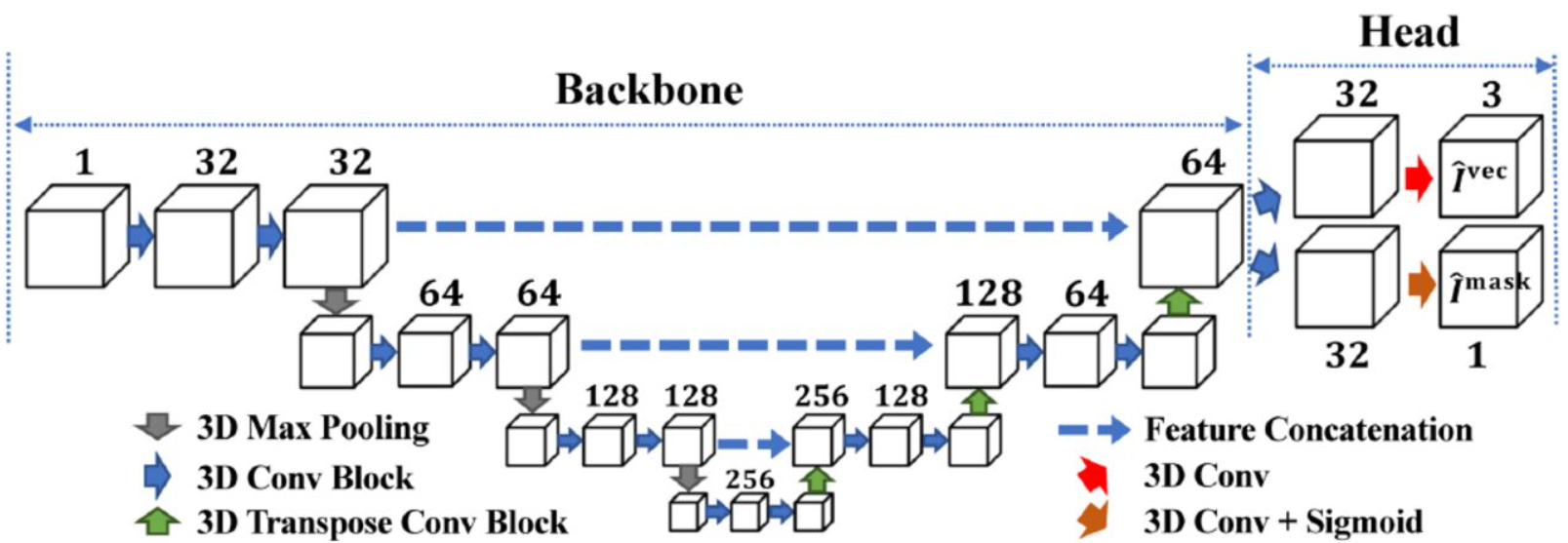
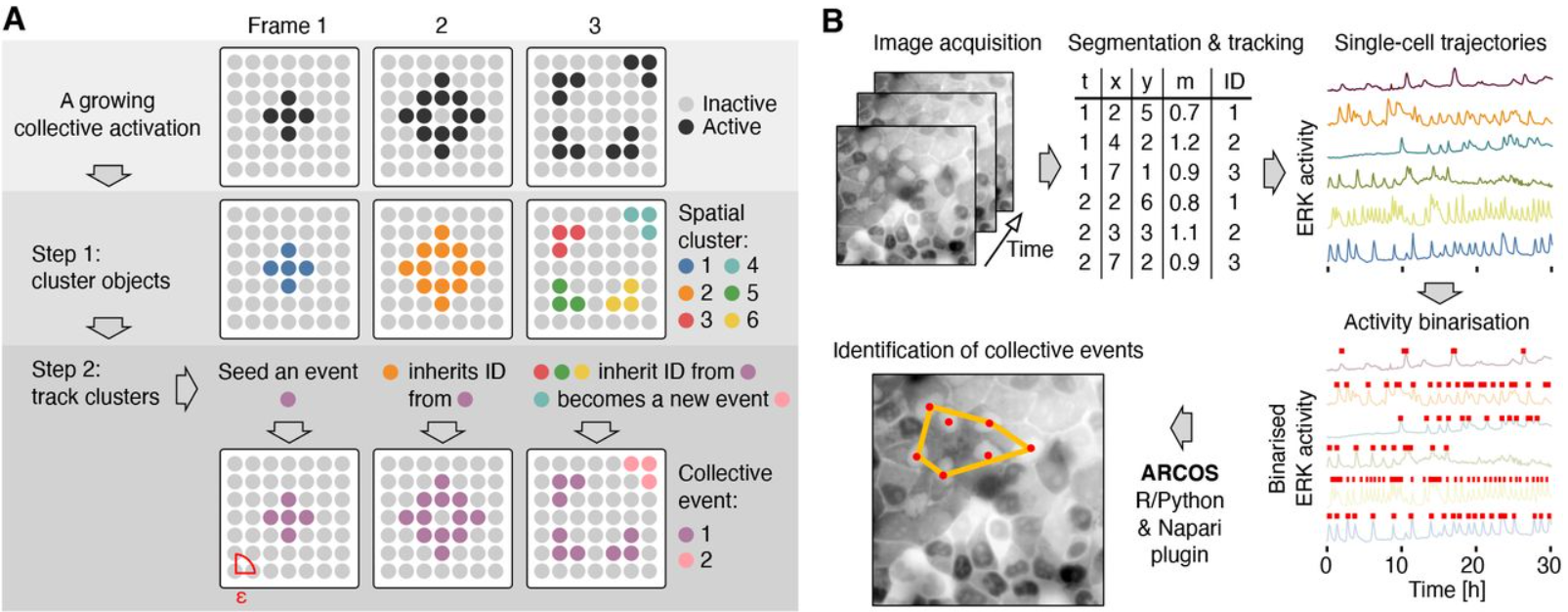
Automatic detection of spatio-temporal signalling patterns in cell collectives
Paolo Armando Gagliardi, Benjamin Grädel, Marc-Antoine Jacques, Lucien Hinderling, Pascal Ender, Andrew R. Cohen, Gerald Kastberger, Olivier Pertz, Maciej Dobrzyński
Efficient End-to-end Learning for Cell Segmentation with Machine Generated Incomplete Annotations
Prem Shrestha, Nicholas Kuang, Ji Yu
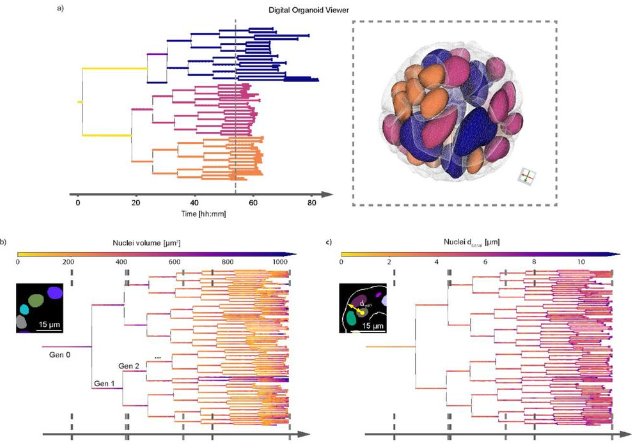
Multiscale light-sheet organoid imaging framework
Gustavo de Medeiros, Raphael Ortiz, Petr Strnad, Andrea Boni, Franziska Moos, Nicole Repina, Ludivine Chalet Meylan, Francisca Maurer, Prisca Liberali
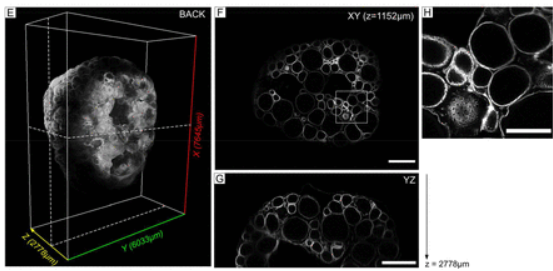
An end-to-end pipeline based on open source deep learning tools for reliable analysis of complex 3D images of Medaka ovaries
Manon Lesage, Jérôme Bugeon, Manon Thomas, Thierry Pécot, Violette Thermes


 (No Ratings Yet)
(No Ratings Yet)