Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 7 October 2022
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools only.
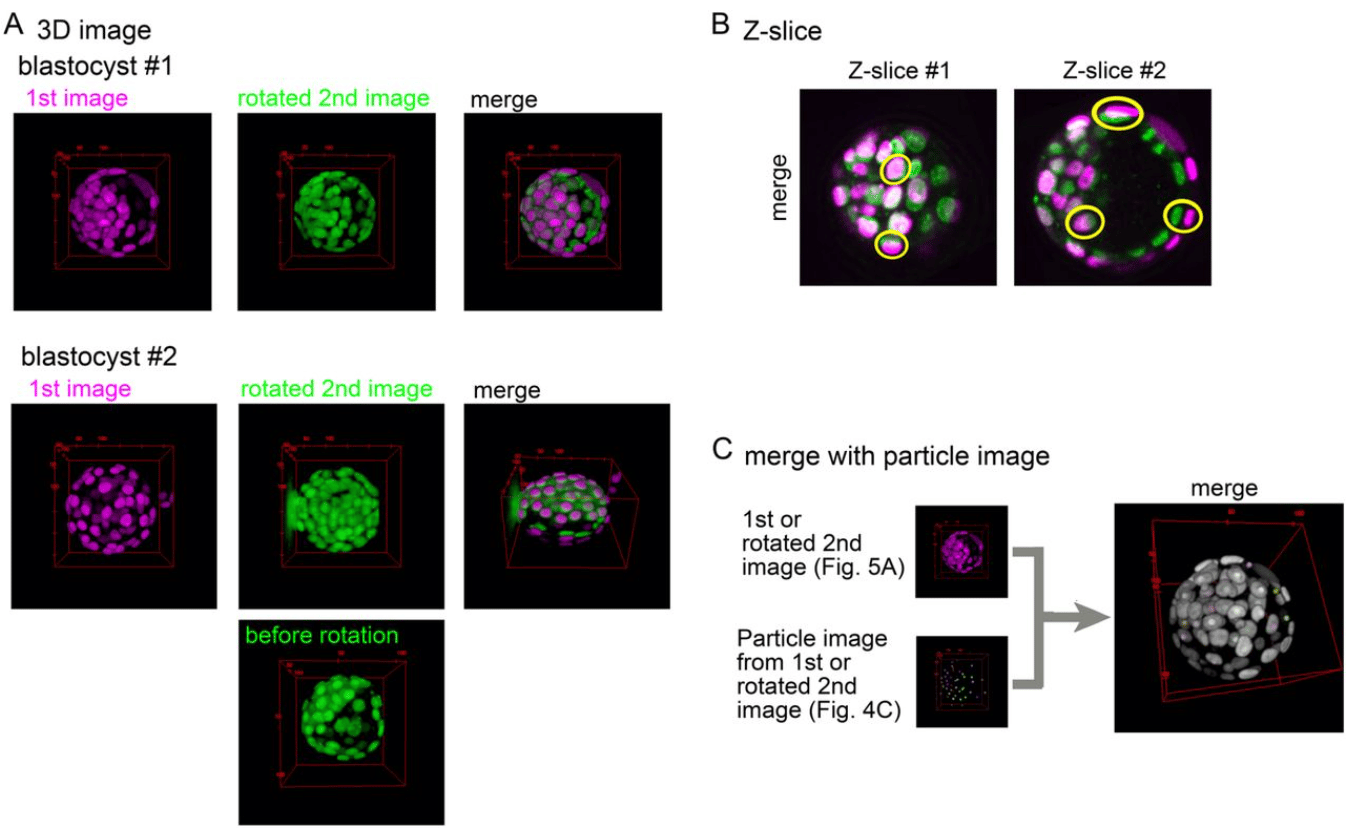
An ImageJ-based tool for three-dimensional registration between different types of microscopic images
Hiroshi Koyama, Kanae Kishi, Toshihiko Fujimori
Quantification of membrane binding and diffusion using Fluorescence Correlation Spectroscopy diffusion laws
Anita Mouttou, Erwan Bremaud, Julien Noero, Rayane Dibsy, Coline Arone, Johnson Mak, Delphine Muriaux, Hugues Berry, Cyril Favard
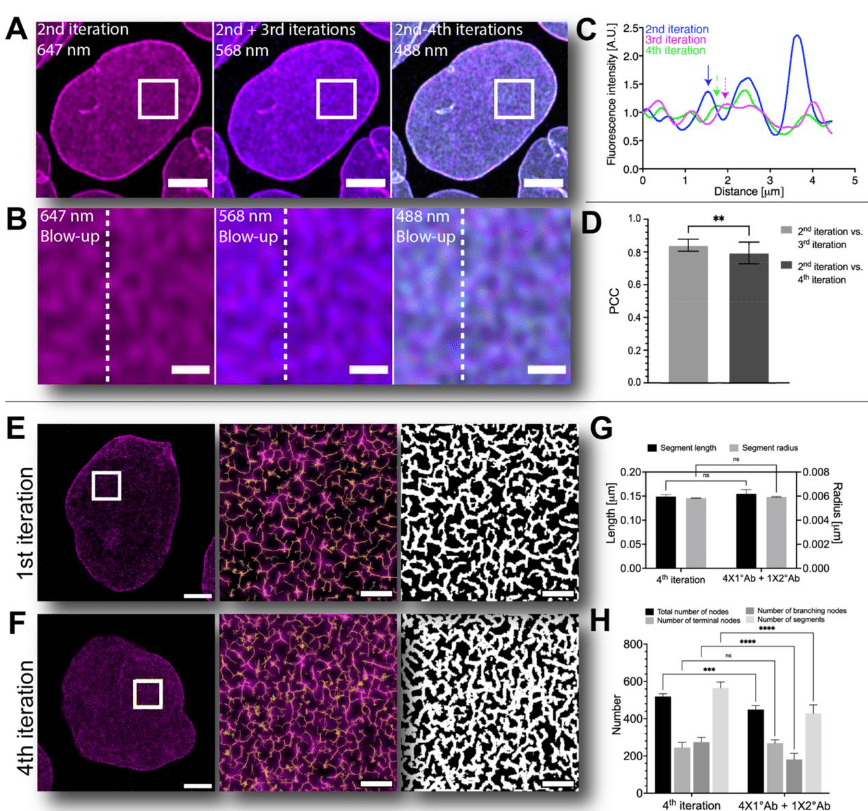
Iterative immunostaining combined with expansion microscopy and image processing reveals nanoscopic network organization of nuclear lamina
Elina Mäntylä, Toni Montonen, Lucio Azzari, Salla Mattola, Markus Hannula, Maija Vihinen-Ranta, Jari Hyttinen, Minnamari Vippola, Alessandro Foi, Soile Nymark, Teemu O. Ihalainen
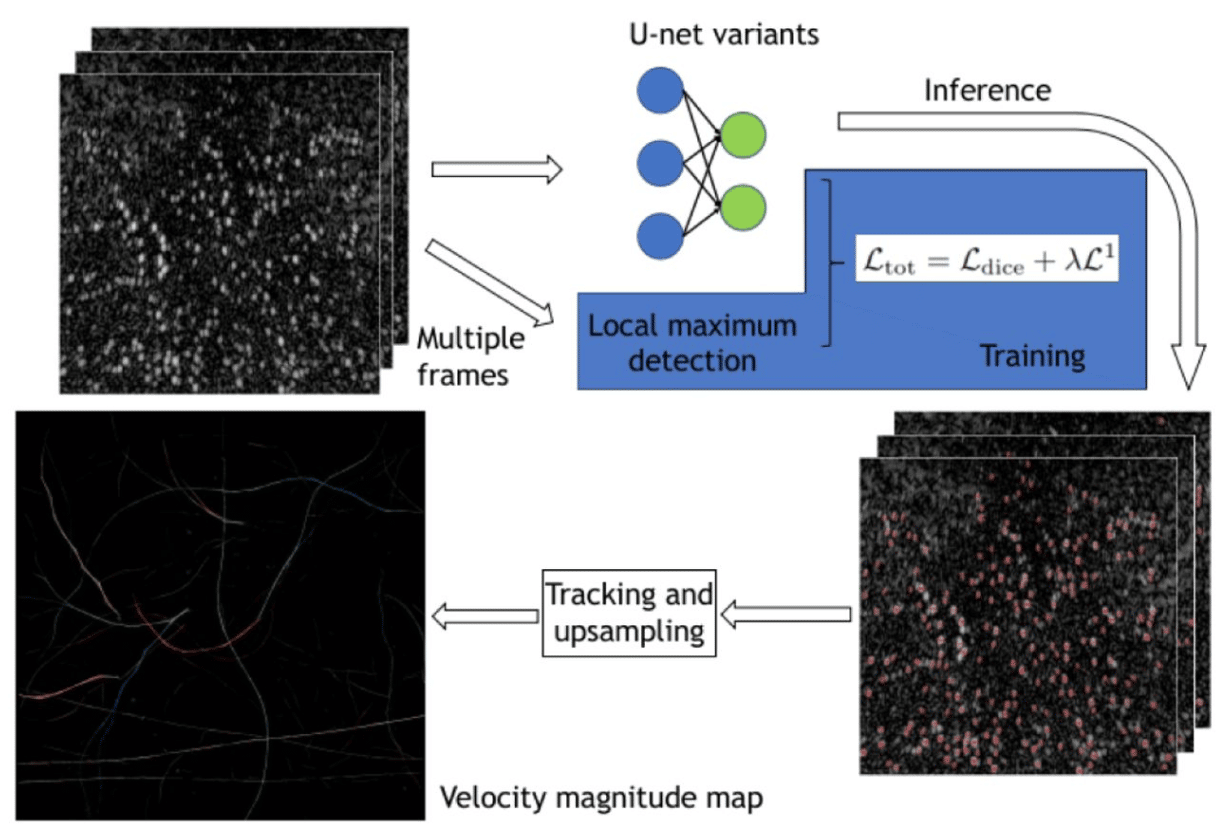
Super resolution ultrasound imaging Using deep learning based micro-bubbles localization
Feixiao Long, Weiguang Zhang
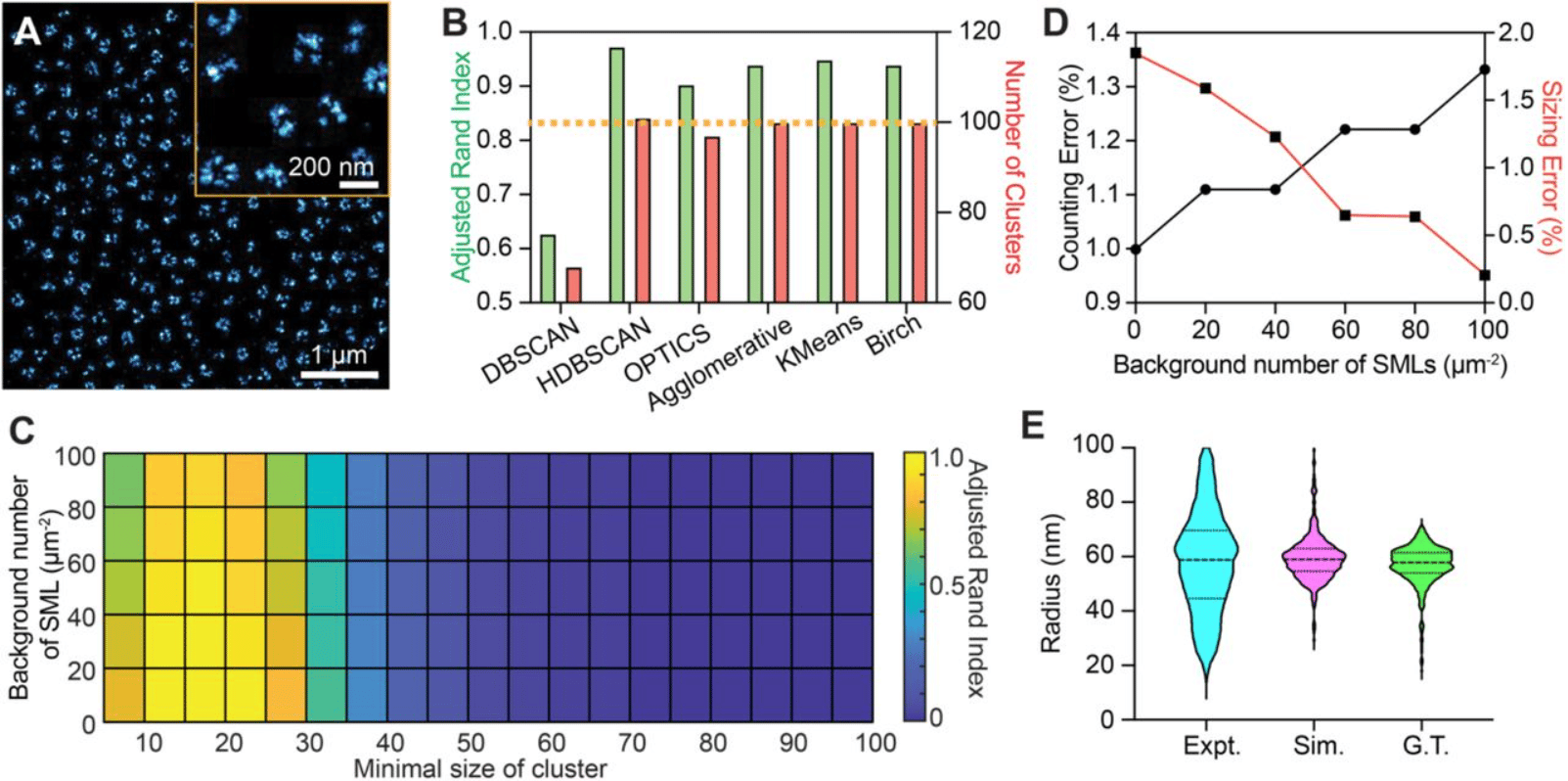
Experimental Parameters-Based Monte-Carlo Simulation of Single-Molecule Localization Microscopy of Nuclear Pore Complex to Evaluate Clustering Algorithms
Wei-Hong Yeo, Yang Zhang, Amy E. Neely, Xiaomin Bao, Cheng Sun, Hao F. Zhang
Gentle and fast all-atom model refinement to cryo-EM densities via Bayes’ approach
Christian Blau, Linnea Yvonnesdotter, Erik Lindahl
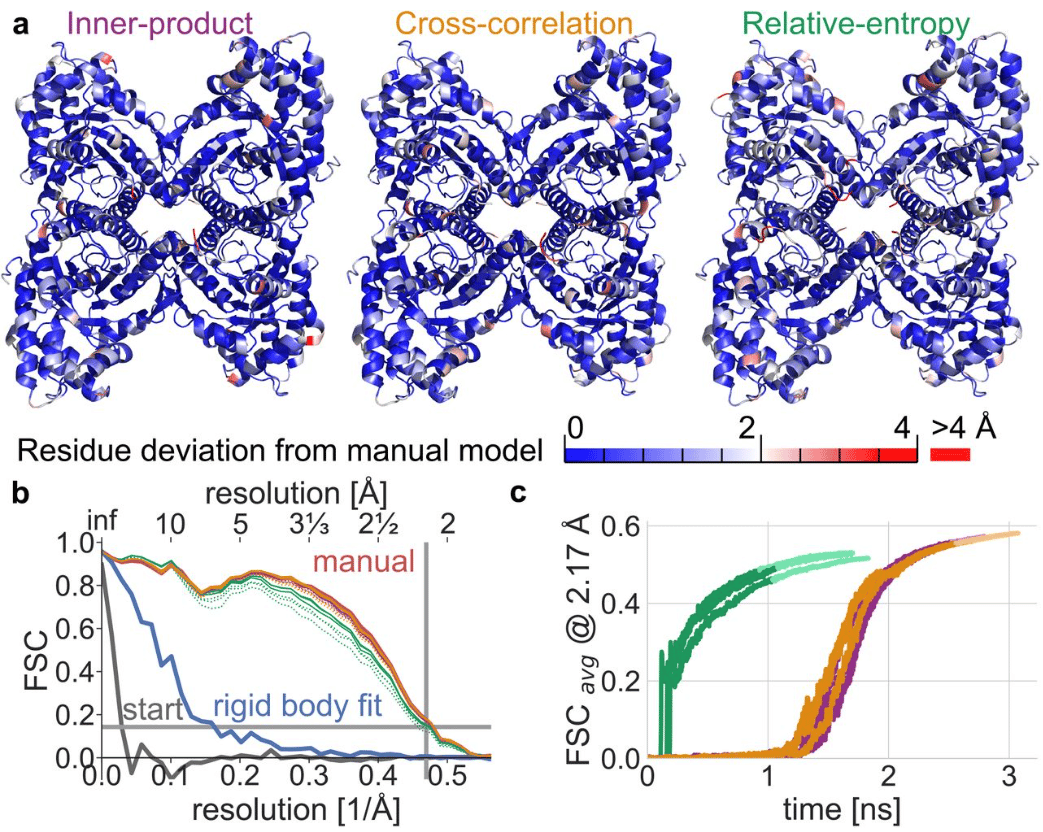
Measuring the similarity of SMLM-derived point-clouds
Mohammed Baragilly, Daniel J. Nieves, David J. Williamson, Ruby Peters, Dylan M. Owen
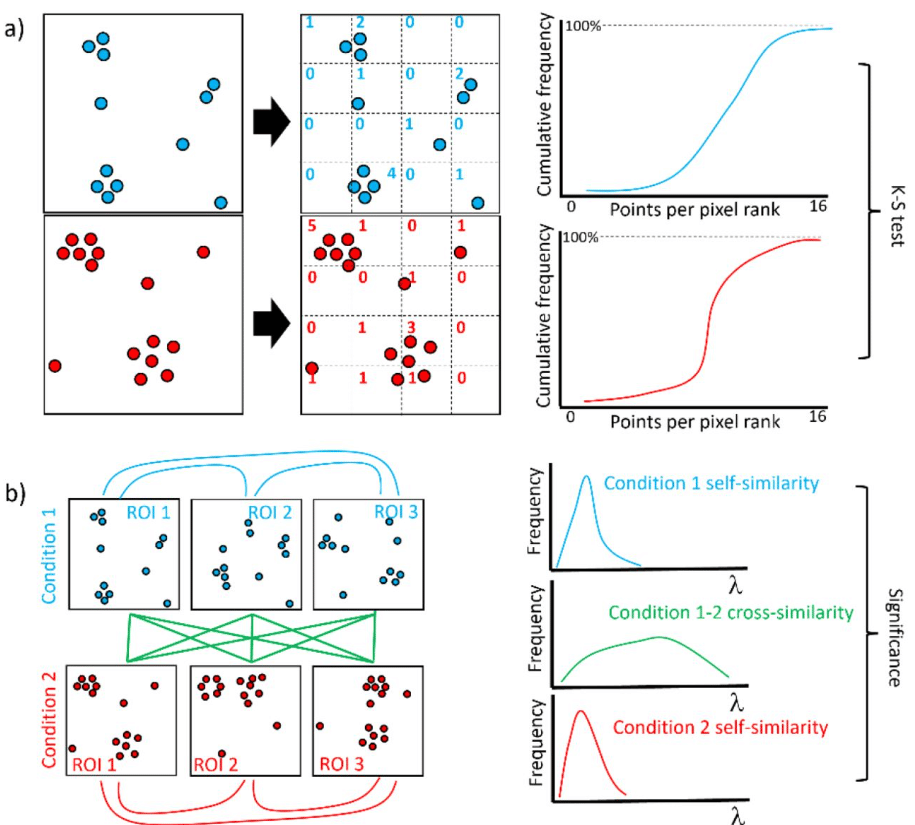
Flexible and open-source programs for quantitative image analysis in microbial ecology
Alexis L. Pasulka, Jonathan F. Hood, Dana E. Michels, Mason D. Wright
TACI: an ImageJ plugin for 3D calcium imaging analysis
Alisa A. Omelchenko, Hua Bai, Sibtain Hussain, Jordan J. Tyrrell, Lina Ni
Rapid and fully automated blood vasculature analysis in 3D light-sheet image volumes of different organs
Philippa Spangenberg, Nina Hagemann, Anthony Squire, Nils Förster, Sascha D. Krauß, Yachao Qi, Ayan Mohamud Yusuf, Jing Wang, Anika Grüneboom, Lennart Kowitz, Sebastian Korste, Matthias Totzeck, Zülal Cibir, Ali Ata Tuz, Vikramjeet Singh, Devon Siemes, Laura Struensee, Daniel R. Engel, Peter Ludewig, Luiza Martins Nascentes Melo, Iris Helfrich, Jianxu Chen, Matthias Gunzer, Dirk M. Hermann, Axel Mosig

Automated image analysis method to detect and quantify fat cell infiltration in hematoxylin and eosin stained human pancreas histology images
Roshan Ratnakar Naik, Annie Rajan, Nehal Kalita
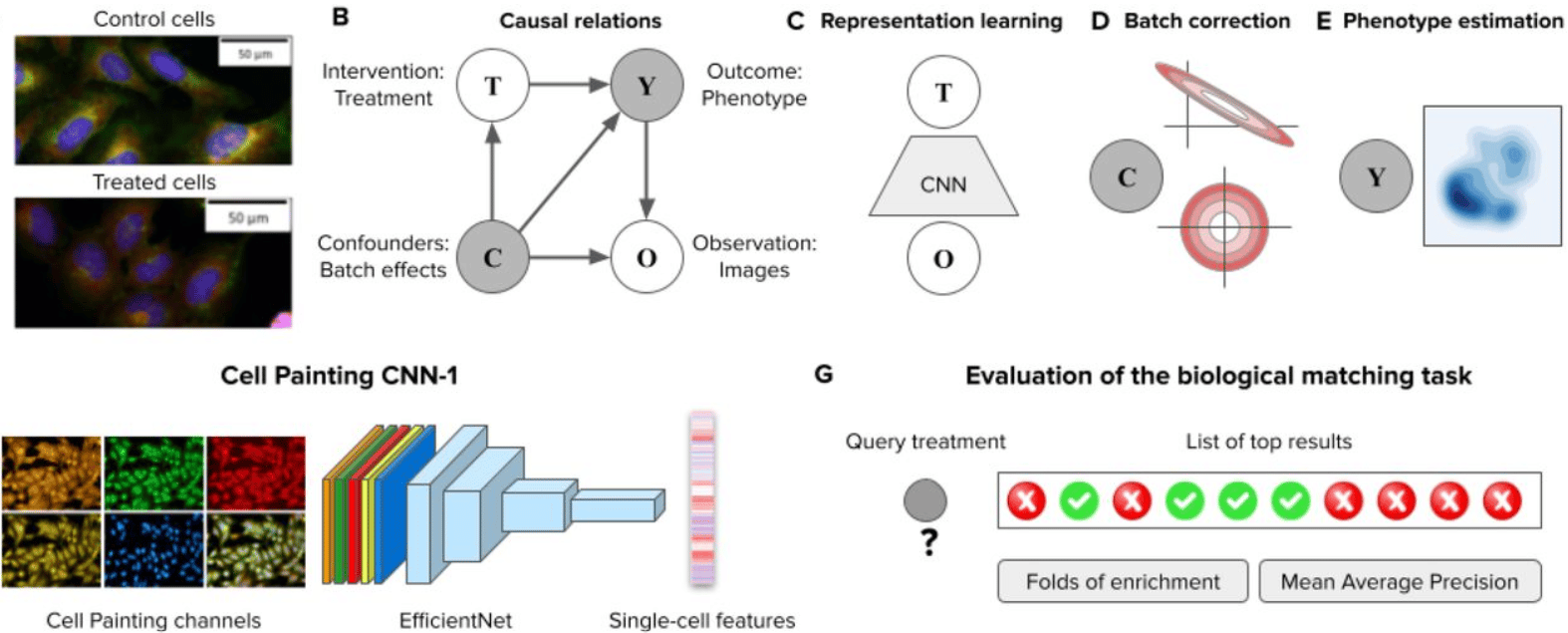
Learning representations for image-based profiling of perturbations
Nikita Moshkov, Michael Bornholdt, Santiago Benoit, Matthew Smith, Claire McQuin, Allen Goodman, Rebecca A. Senft, Yu Han, Mehrtash Babadi, Peter Horvath, Beth A. Cimini, Anne E. Carpenter, Shantanu Singh, Juan C. Caicedo
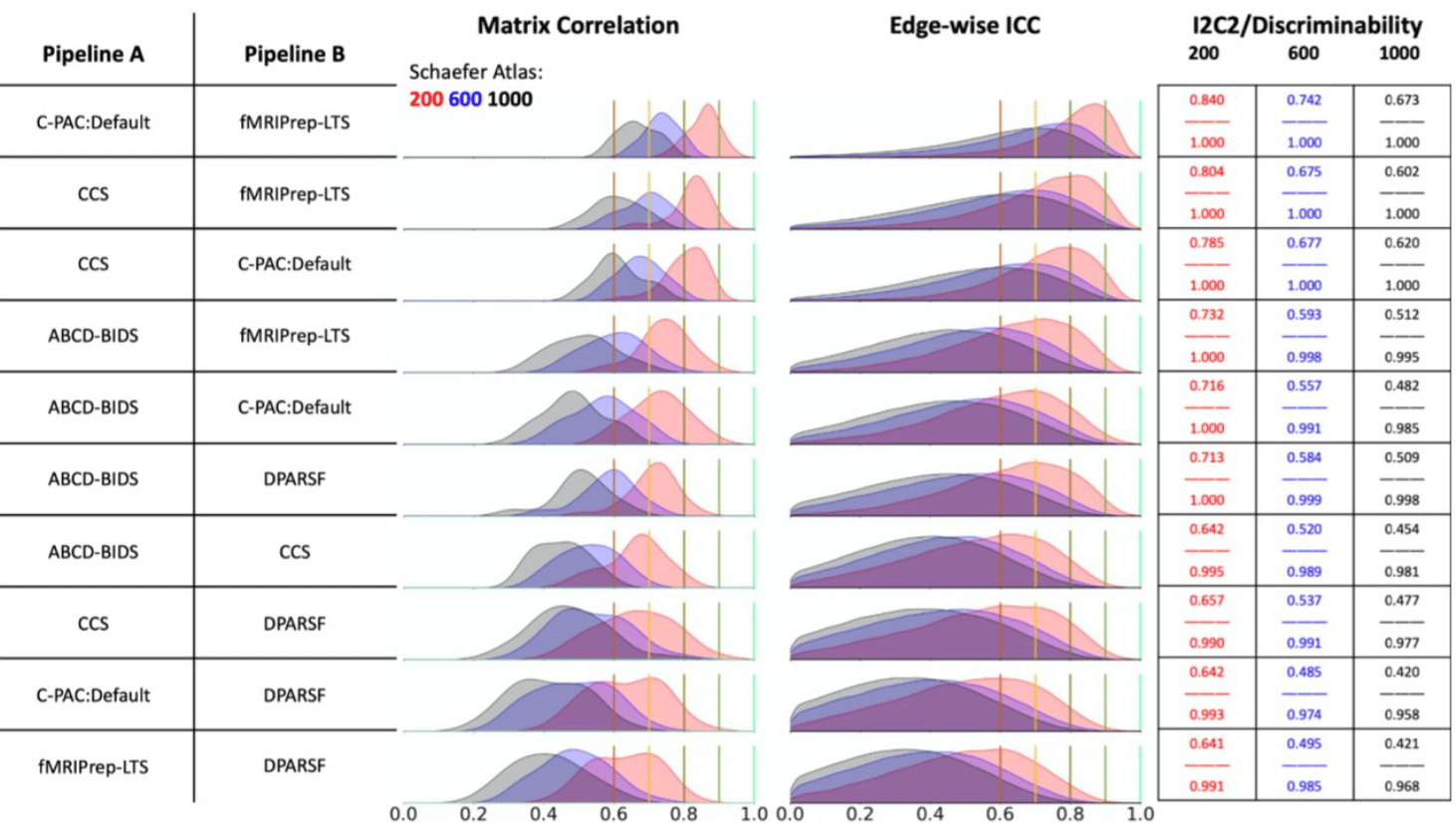
Moving Beyond Processing and Analysis-Related Variation in Neuroscience
Xinhui Li, Lei Ai, Steve Giavasis, Hecheng Jin, Eric Feczko, Ting Xu, Jon Clucas, Alexandre Franco, Anibal Sólon Heinsfeld, Azeez Adebimpe, Joshua T. Vogelstein, Chao-Gan Yan, Oscar Esteban, Russell A. Poldrack, Cameron Craddock, Damien Fair, Theodore Satterthwaite, Gregory Kiar, Michael P. Milham
High-throughput cell spheroid production and assembly analysis by microfluidics and deep learning
Martin Trossbach, Emma Akerlund, Krzysztof Langer, Brinton Seashore-Ludlow, Haakan N. Joensson


 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)