Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 18 January 2023
Here is a curated selection of preprints published recently. In this post, we we focus specifically on new bioimage analysis tools.
AimSeg: a machine-learning-aided tool for axon, inner tongue and myelin segmentation
Ana Maria Rondelli, Jose Manuel Morante-Redolat, Peter Bankhead, Bertrand Vernay, Anna Williams, Pau Carrillo-Barberà
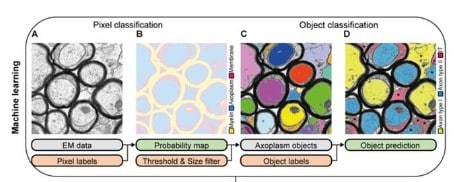
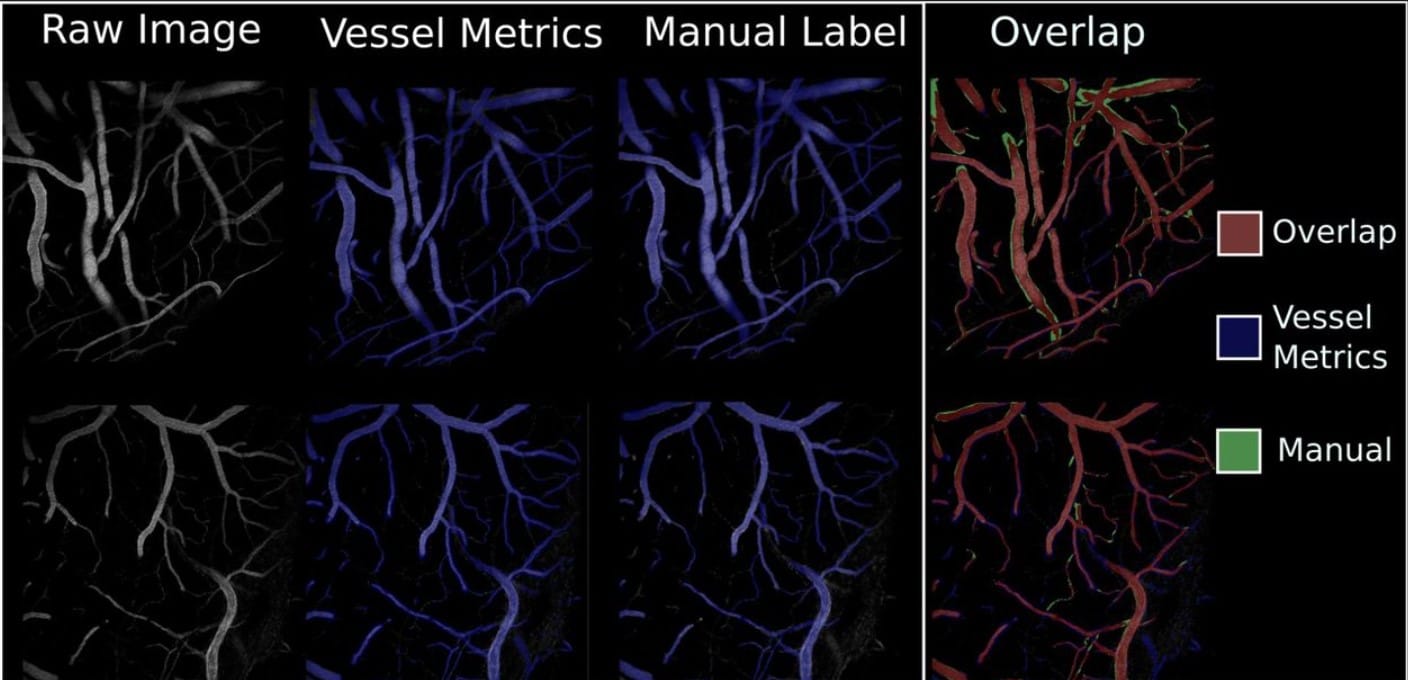
Vessel Metrics: A python based software tool for automated analysis of vascular structure in confocal imaging
Sean D. McGarry, Cynthia Adjekukor, Suchit Ahuja, Jasper Greysson-Wong, Idy Vien, Kristina D. Rinker, Sarah.J. Childs
pomBseen: An Automated Pipeline for Analysis of Fission Yeast Images
Makoto Ohira, Nicholas Rhind
Sketch the Organoids from Birth to Death – Development of an Intelligent OrgaTracker System for Multi-Dimensional Organoid Analysis and Recreation
Xuan Du, Wenhao Cui, Jiaping Song, Yanping Cheng, Yuxin Qi, Yue Zhang, Qiwei Li, Jing Zhang, Lifeng Sha, Jianjun Ge, Yanhui Li, Zaozao Chen, Zhongze Gu
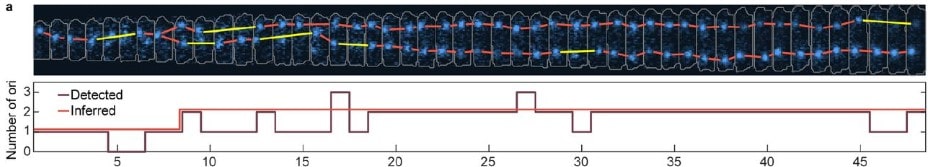
★Track: Inferred counting and tracking of replicating DNA loci
Robin Köhler, Ismath Sadhir, Seán M. Murray
A deep learning-based stripe self-correction method for stitched microscopic images
Shu Wang, Xiaoxiang Liu, Yueying Li, Xinquan Sun, Qi Li, Yinhua She, Yixuan Xu, Xingxin Huang, Ruolan Lin, Deyong Kang, Xingfu Wang, Haohua Tu, Wenxi Liu, Feng Huang, Jianxin Chen
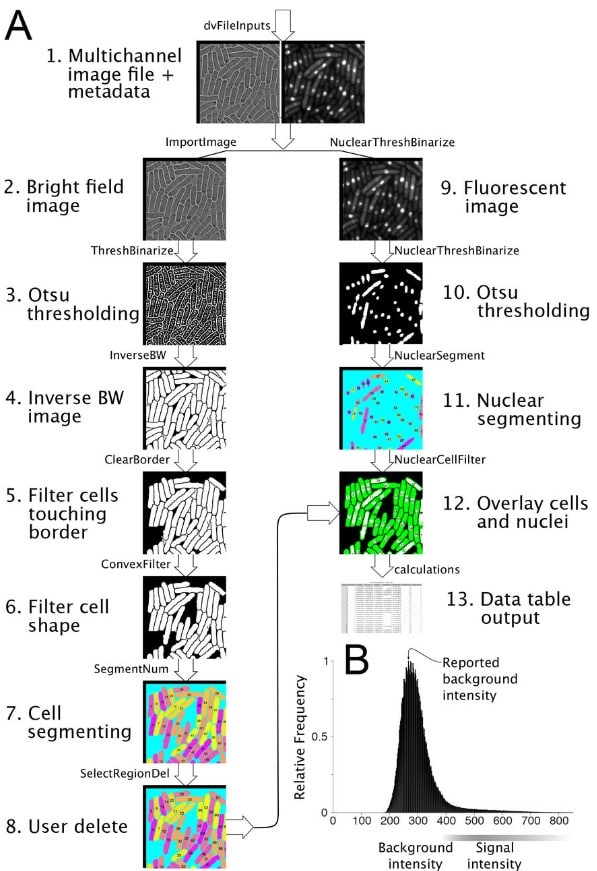
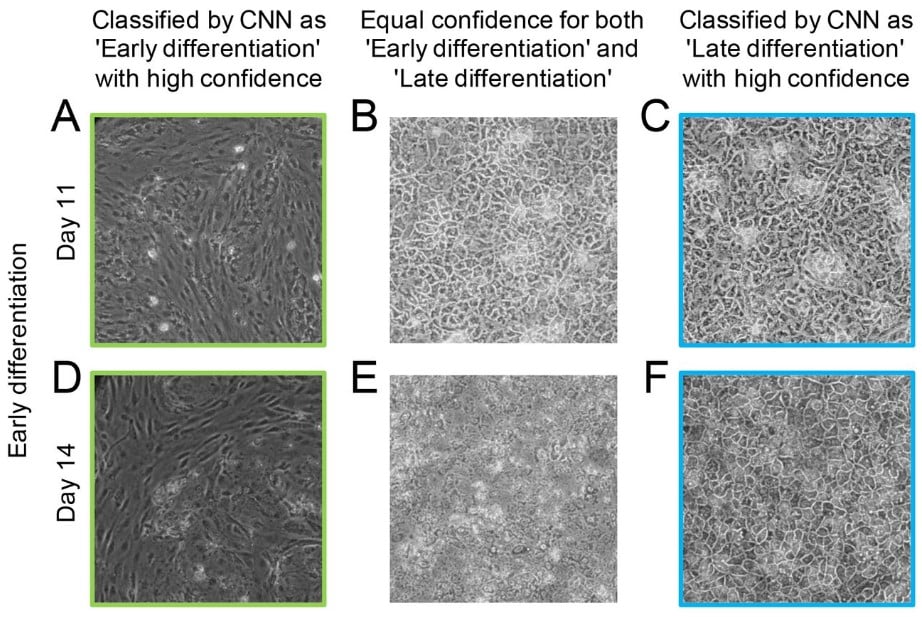
Artificial intelligence supports automated characterization of differentiated human pluripotent stem cells
Katarzyna Marzec-Schmidt, Nidal Ghosheh, Sören Richard Stahlschmidt, Barbara Küppers-Munther, Jane Synnergren, Benjamin Ulfenborg
Quantitatively mapping local quality of super-resolution microscopy by rolling Fourier ring correlation
Weisong Zhao, Xiaoshuai Huang, Jianyu Yang, Guohua Qiu, Liying Qu, Yue Zhao, Shiqun Zhao, Ziying Luo, Xinwei Wang, Yaming Jiu, Heng Mao, Xumin Ding, Jiubin Tan, Ying Hu, Leiting Pan, Liangyi Chen, Haoyu Li
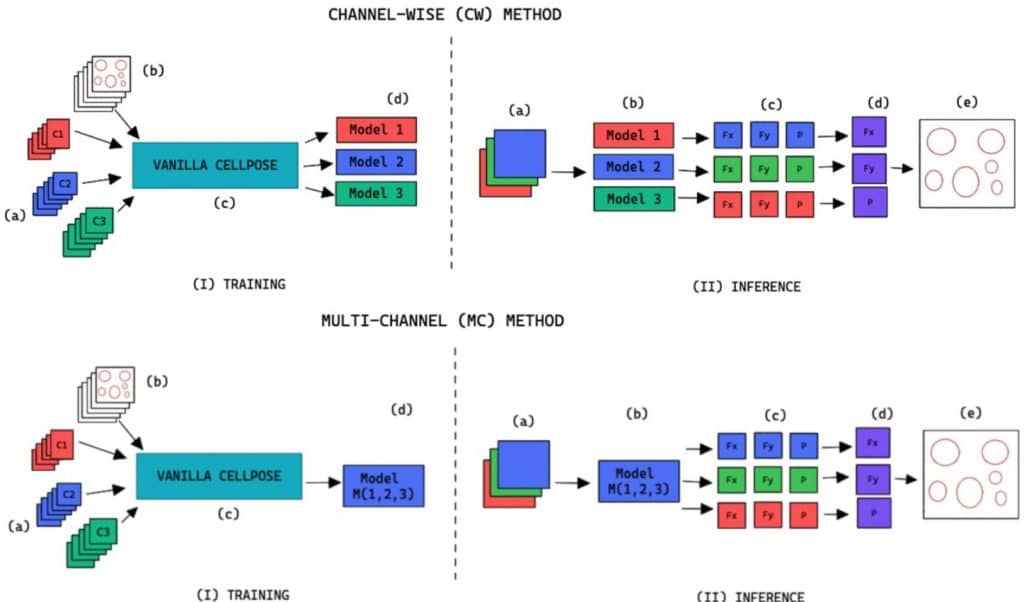
Cell Segmentation in Images Without Structural Fluorescent Labels
Daniel Zyss, Susana A. Ribeiro, Mary J.C. Ludlam, Thomas Walter, Amin Fehri
A practical extraction and spatial statistical pipeline for large 3D bioimages
George Adams, Floriane Tissot, Chang Liu, Chris Brunsdon, Ken R. Duffy, Cristina Lo Celso
Unmixing Biological Fluorescence Image Data with Sparse and Low-Rank Poisson Regression
Ruogu Wang, Alex A. Lemus, Colin M. Henneberry, Yiming Ying, Yunlong Feng, Alex M. Valm
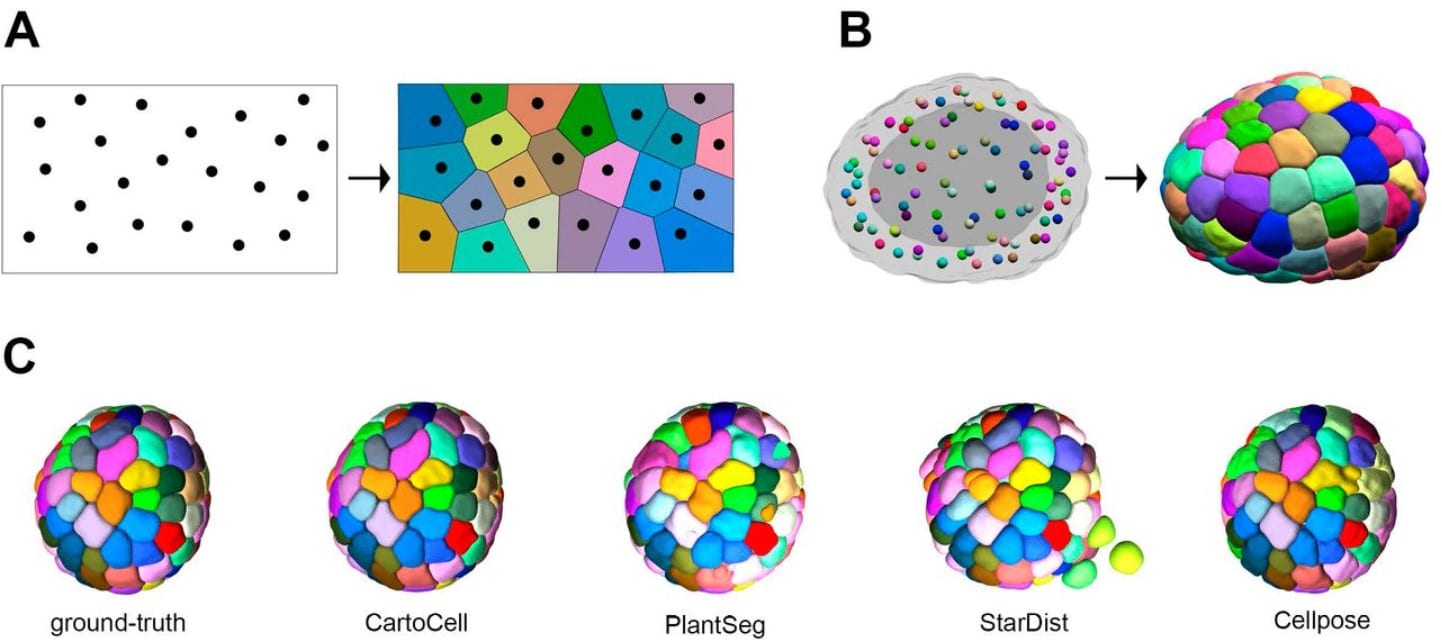
CartoCell, a high-throughput pipeline for accurate 3D image analysis, unveils cell morphology patterns in epithelial cysts
Jesús A. Andrés-San Román, Carmen Gordillo-Vázquez, Daniel Franco-Barranco, Laura Morato, Antonio Tagua, Pablo Vicente-Munuera, Ana M. Palacios, María P. Gavilán, Valentina Annese, Pedro Gómez-Gálvez, Ignacio Arganda-Carreras, Luis M. Escudero
Weakly-supervised temporal segmentation of cell-cycle stages with center-cell focus using Recurrent Neural Networks
Abin Jose, Rijo Roy, Johannes Stegmaier
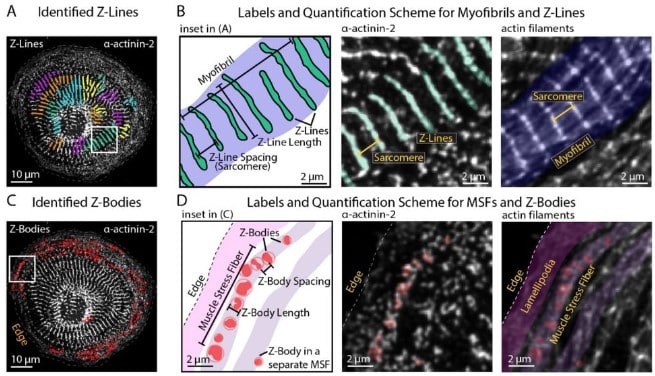
Independent regulation of Z-lines and M-lines during sarcomere assembly in cardiac myocytes revealed by the automatic image analysis software sarcApp
Abigail C. Neininger-Castro, James B. Hayes Jr., Zachary C. Sanchez, Nilay Taneja, Aidan M. Fenix, Satish Moparthi, Stéphane Vassilopoulos, Dylan T. Burnette
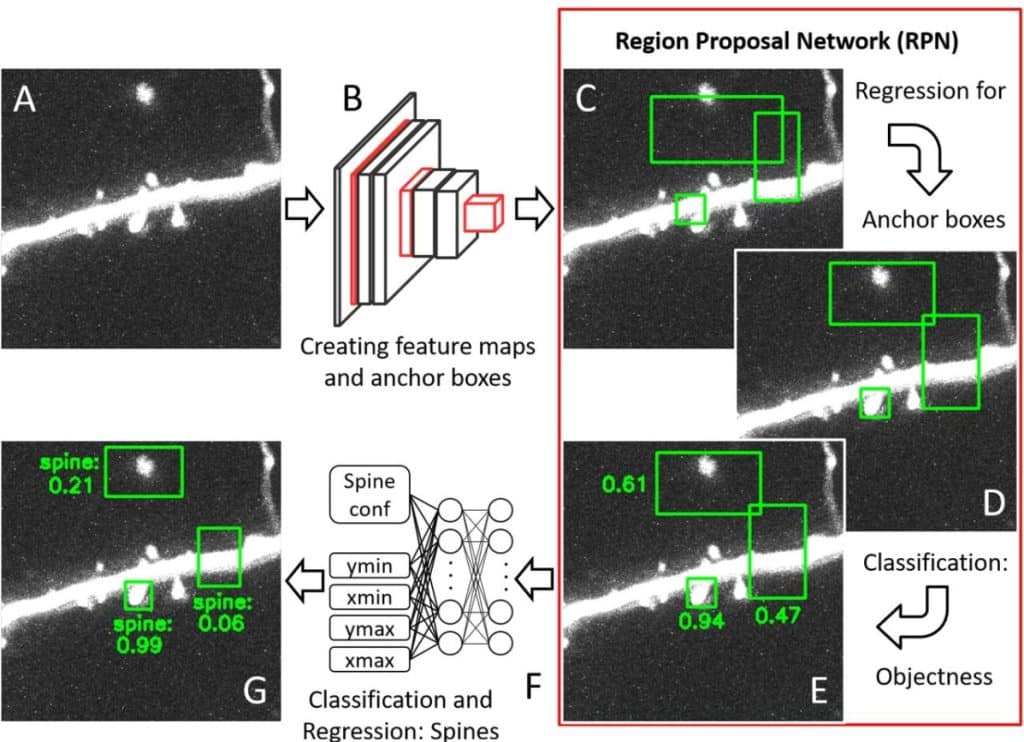
Fully automated detection of dendritic spines in 3D live cell imaging data using deep convolutional neural networks
Fabian W. Vogel, Sercan Alipek, Jens-Bastian Eppler, Jochen Triesch, Diane Bissen, Amparo Acker-Palmer, Simon Rumpel, Matthias Kaschube
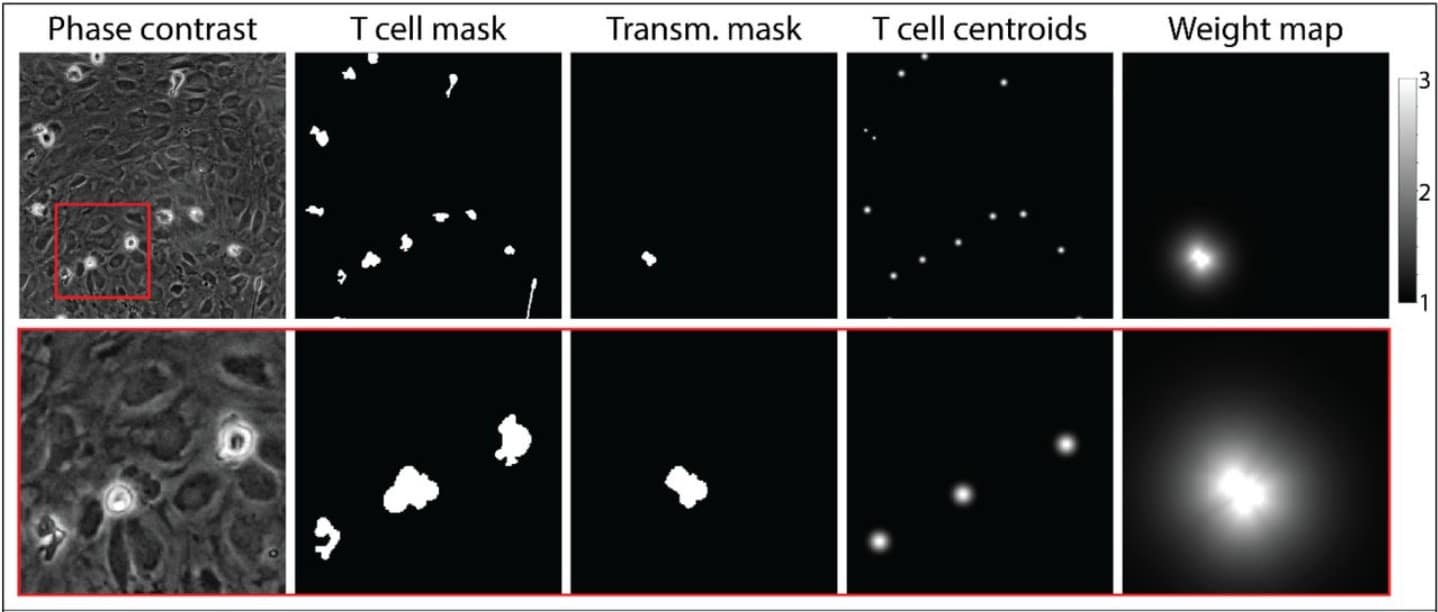
UFMTrack: Under-Flow Migration Tracker enabling analysis of the entire multi-step immune cell extravasation cascade across the blood-brain barrier in microfluidic devices
Mykhailo Vladymyrov, Luca Marchetti, Sidar Aydin, Sasha Soldati, Adrien Mossu, Arindam Pal, Laurent Gueissaz, Akitaka Ariga, Britta Engelhardt
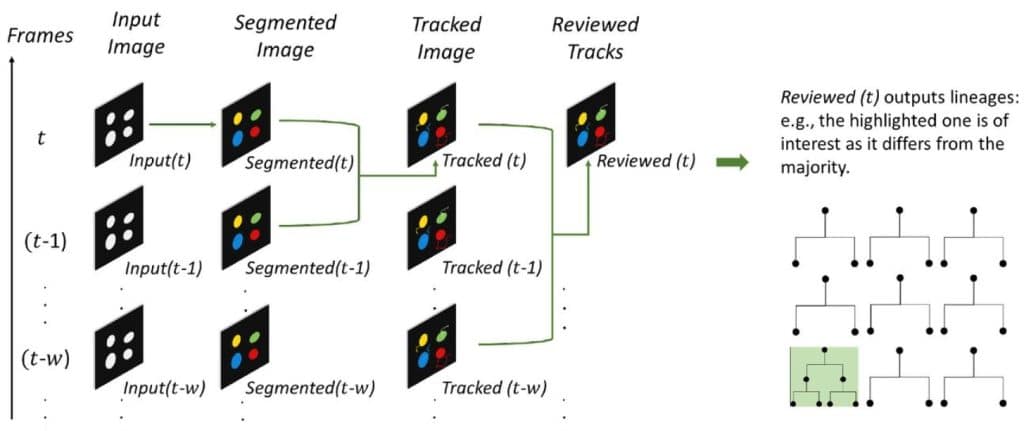
Fast and Accurate Cell Tracking: a real-time cell segmentation and tracking algorithm to instantly export quantifiable cellular characteristics from large scale image data
Ting-Chun Chou, Li You, Cecile Beerens, Kate J. Feller, Miao-Ping Chien
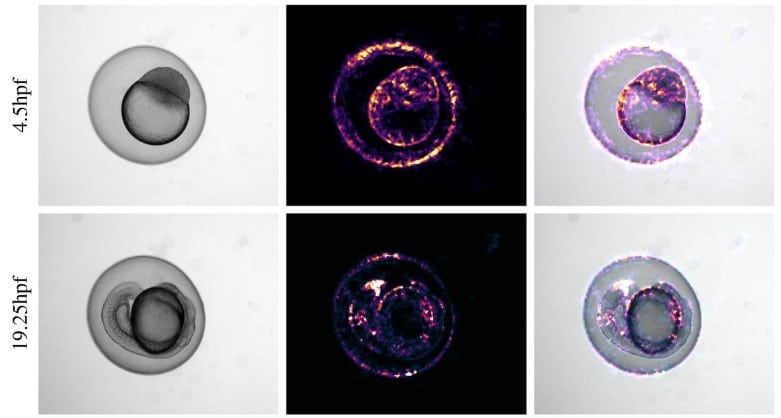
Automated staging of zebrafish embryos with KimmelNet
David J. Barry, Rebecca A. Jones, Matthew J. Renshaw


 (No Ratings Yet)
(No Ratings Yet)
this is extremely helpful. is it possible to continue the curation? I can contribute