Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 1 March 2023
Here is a curated selection of preprints published recently. In this post, we focus specifically on new bioimage analysis tools, including an introduction to quantification of microscopy data from Siân Culley and colleagues.
Zero-shot learning enables instant denoising and super-resolution in optical fluorescence microscopy
Chang Qiao, Yunmin Zeng, Quan Meng, Xingye Chen, Haoyu Chen, Tao Jiang, Rongfei Wei, Jiabao Guo, Wenfeng Fu, Huaide Lu, Di Li, Yuwang wang, Hui Qiao, Jiamin Wu, Dong Li, Qionghai Dai
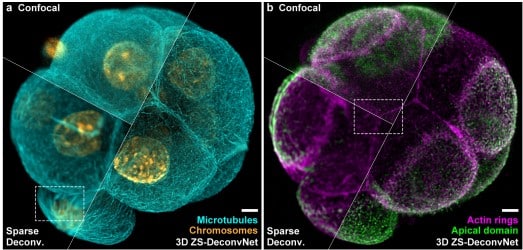
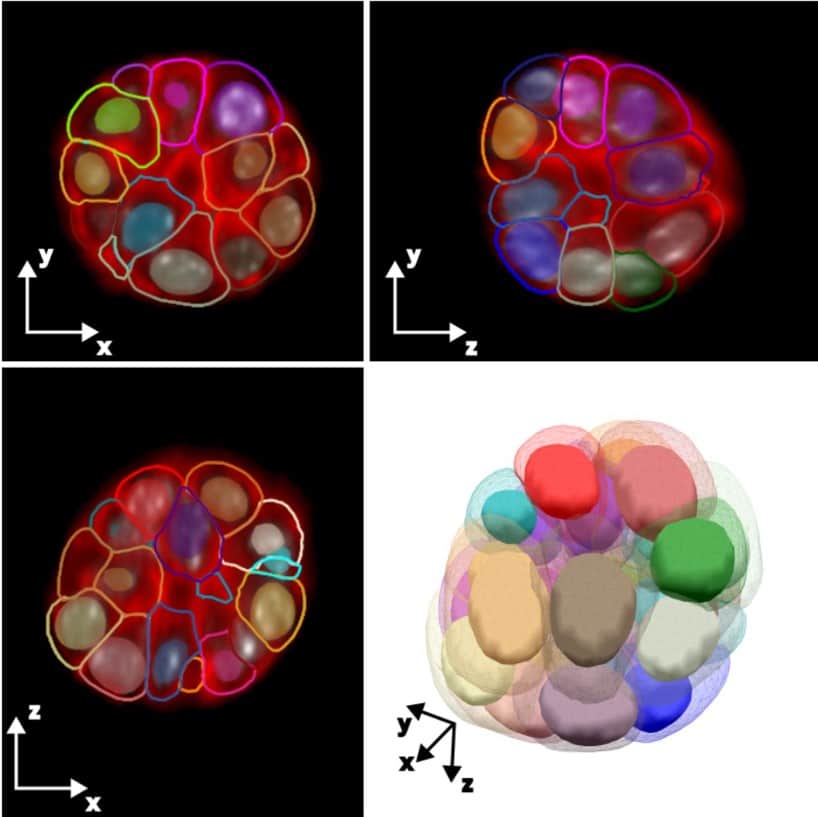
Active mesh and neural network pipeline for cell aggregate segmentation
Matthew B. Smith, Hugh Sparks, Jorge Almagro, Agathe Chaigne, Axel Behrens, Chris Dunsby, Guillaume Salbreux
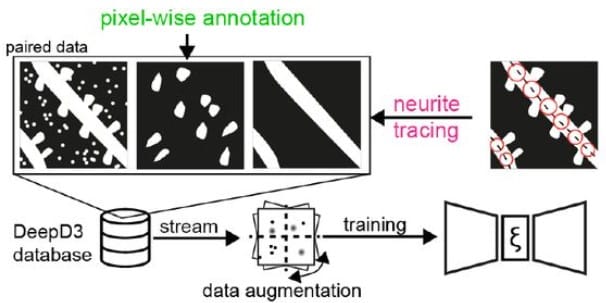
DeepD3, an Open Framework for Automated Quantification of Dendritic Spines
Martin H P Fernholz, Drago A Guggiana Nilo, Tobias Bonhoeffer, Andreas M Kist
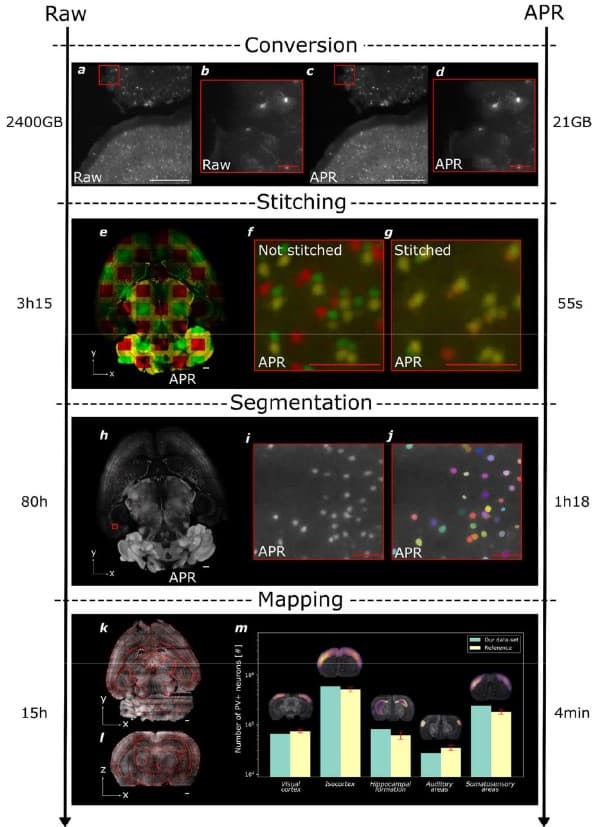
Efficient image analysis for large-scale next generation histopathology using pAPRica
Jules Scholler, Joel Jonsson, Tomás Jordá-Siquier, Ivana Gantar, Laura Batti, Bevan L. Cheeseman, Stéphane Pagès, Ivo F. Sbalzarini, Christophe M. Lamy
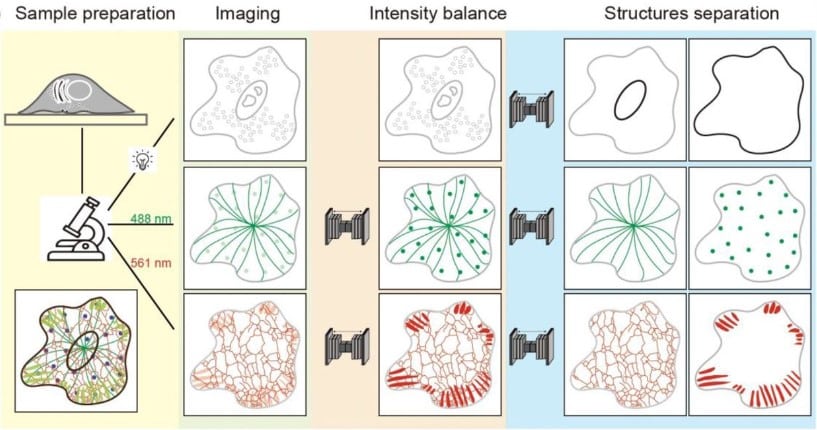
Artificial channel separation of monochrome multi-structure microscopy image
Luhong Jin, Jingfang Liu, Heng Zhang, Yunqi Zhu, Haixu Yang, Jianhang Wang, Luhao Zhang, Yingke Xu
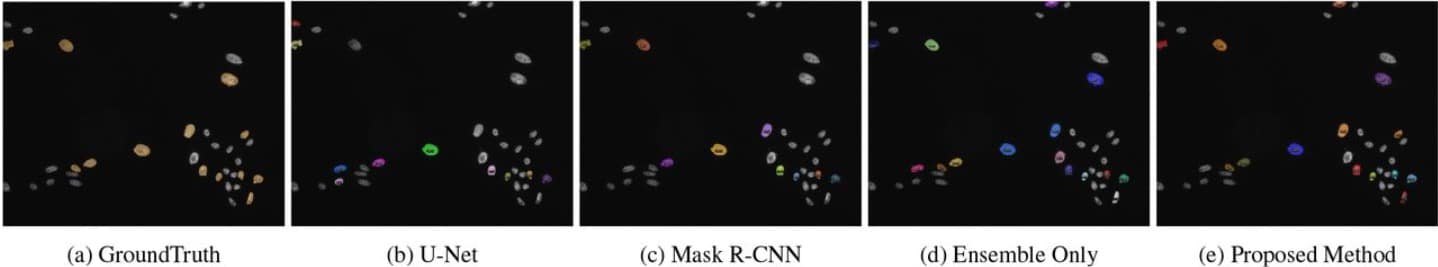
Ensemble Processing and Synthetic Image Generation for Abnormally Shaped Nuclei Segmentation
Yue Han, Yang Lei, Viktor Shkolnikov, Daisy Xin, Alicia Auduong, Steven Barcelo, Jan Allebach, Edward J. Delp
Deep-Learning Assisted, Single-molecule Imaging analysis (Deep-LASI) of multi-color DNA Origami structures
Simon Wanninger, Pooyeh Asadiatouei, Johann Bohlen, Clemens-Bässem Salem, Philip Tinnefeld, Evelyn Ploetz, Don C. Lamb
A simple image processing pipeline to sharpen topology maps in multi-wavelength interference microscopy
Peter W. Tinning, Jana K. Schniete, Ross Scrimgeour, Lisa S. Kölln, Liam M. Rooney, Trevor J. Bushell, Gail McConnell
Label-Free Mammalian Cell Tracking Enhanced by Precomputed Velocity Fields
Yue Han, Yang Lei, Viktor Shkolnikov, Daisy Xin, Steven Barcelo, Jan Allebach, Edward J. Delp
FriendlyClearMap: An optimized toolkit for mouse brain mapping and analysis
Moritz Negwer, Bram Bosch, Maren Bormann, Rick Hesen, Lukas Lütje, Lynn Aarts, Carleen Rossing, Nael Nadif Kasri, Dirk Schubert
Small Training Dataset Convolutional Neural Networks for Application Specific Super-Resolution Microscopy
Varun Mannam, Scott Howard
Quantifying yeast microtubules and spindles using the Toolkit for Automated Microtubule Tracking (TAMiT)
Saad Ansari, Zachary R. Gergely, Patrick Flynn, Gabriella Li, Jeffrey K. Moore, Meredith D. Betterton
Made to measure: an introduction to quantification in microscopy data
Siân Culley, Alicia Cuber Caballero, Jemima J Burden, Virginie Uhlmann
Culley and colleagues


 (No Ratings Yet)
(No Ratings Yet)