Leveraging biophysics to push the limits of investigating RNAs in cells: Riboglow-FLIM, a breakthrough tool for visualizing RNAs in live mammalian cells
Posted by nadia sarfraz, on 1 May 2023
Nadia Sarfraz1, Esther Braselmann1*
1 Department of Chemistry, Georgetown University, Washington, District of Columbia, USA
*corresponding author
Ribonucleic acids (RNAs) play a critical role in cellular function. Visualizing RNAs in space and time in the context of living cells is critical to advance our understanding of biological processes in health and disease.1 Recently, our team developed a tool to do just that called Riboglow-FLIM.2 Riboglow-FLIM provides an innovative way to visualize RNAs and quantify subcellular dynamics. Noteworthy, this platform overcomes notorious artifacts observed from intensity-based imaging platforms, where concentration dependency leads to extreme cell-to-cell heterogeneity. Similarly, intensity-based imaging has inherent limitations that limit multiplexing capabilities, as observed for other RNA labeling systems (e.g., RhoBAST, Spinach, Broccoli, Riboglow version 1, MS2, Mango, Peppers, o-Choral/Gemini-561 and SRB/DNB).3–13
Riboglow-FLIM is a robust platform for multiplexed sensing of different RNAs simultaneously in live mammalian cells using fluorescence lifetime imaging microscopy (FLIM).2 Riboglow-FLIM builds upon “Riboglow version 1” by incorporating an additional dimension of analysis, namely the fluorescence lifetime of the fluorophore component in the small molecule ligand of Riboglow.2,13 The parent Riboglow system involves genetically tagging RNAs with a bacterial riboswitch and adding a small molecular probe (such as Cbl-4xGly-ATTO 590) that undergoes fluorescence turn-on upon binding.13 The bacterial riboswitch is an optimal RNA tag due to its robust folding and specific and tight binding to its natural ligand (Cobalamin, Cbl). Cbl is part of the small molecule probe.14 Importantly, Cbl also has quenching properties; hence the fluorescence of the fluorophore in the probe is reduced, and fluorescence increases upon binding to the RNA, likely due to steric separation of Cbl and fluorophore in the RNA/probe complex.
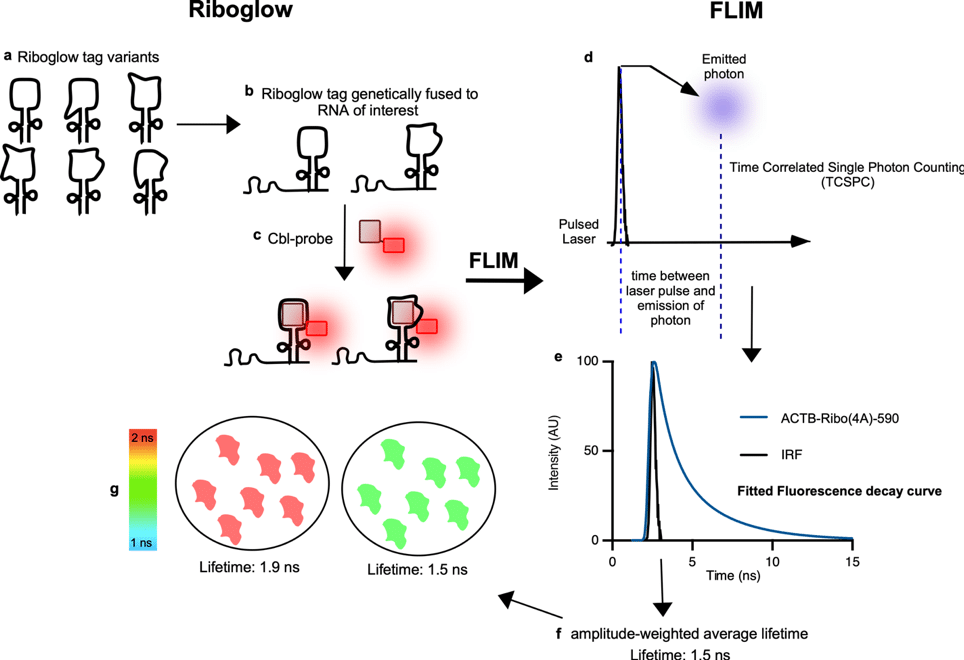
The general principle of Riboglow-FLIM (Figure 1) is that the fluorescence lifetime of each pixel is measured by laser scanning microscopy using Time-Correlated Single Photon Counting (TCSPC), where photon arrival times at the detector after pulsed laser excitation are recorded.2 The acquired decay function is then analyzed using n-exponential re-convolution fitting. We analyzed live mammalian cells in this way for different conditions, for example by comparing cells in the presence of genetically encoded RNA reporters and the chemical probe versus cells that contained the probe, but not the RNA tag.2 The resulting amplitude-weighted average lifetime is then assigned to a pseudo-color scale for visualization. All respective data is extracted and fitted using Symphotime64 re-convolution global analysis (Picoquant).2 For each region of interest (ROI, for example, a whole cell), an average lifetime value can be computed (Figure 1g).
In designing this study, we hypothesized that visualizing Riboglow by FLIM (Riboglow-FLIM) would be advantageous for robust contrast in RNA sensing. We began by comparing the fluorescence lifetime of the probe Cbl-4xGly-ATTO 590 in the presence of two RNA tag variant that we purified, called A and D and observed robust differences.2 We then chose mRNA that encodes for β-actin (ACTB) as a model mRNA to establish Riboglow-FLIM in live mammalian cells, as this system was previously used to evaluate Riboglow performance.2,13 We established a FLIM workflow for Riboglow-FLIM visualization in live mammalian cells by performing multiexponential re-convolution fitting and analyzing whole cells as an ROI.2 We compared cells that produced ACTB-Ribo(4D)-590 versus control cells that were not transfected with the reporter, and only the Riboglow probe was loaded into cells. Importantly, the contrast between cells that produce the Riboglow reporter and untransfected control cells was much greater for FLIM visualization versus intensity-based imaging.2 We also assessed whether the FLIM signal distribution for Riboglow-tagged mRNA accurately reports on subcellular mRNA localization. For this, we used the phenotype of RNA-containing stress granules as a tractable localization pattern and confirmed subcellular localizations with Riboglow-FLIM live that were established in fixed cells previously. Together, we observed a robust increase in fluorescence lifetime for Riboglow-tagged RNA versus the probe alone.2
The use of FLIM has many key advantages, most importantly the quantitative nature of extracted data. On the other hand, the data collection and analysis is not trivial. To make this approach accessible, we share “Pro-tips” here.
- When using Riboglow-FLIM, it is crucial to consider the health and time frame of imaging, particularly when working with live cells. Based on our findings, cells imaged beyond the suggested time frame of approximately 2-3 hours, or when cells are not maintained in optimal cellular conditions, may exhibit erroneous lifetimes. This underscores the importance of meticulous control over the cellular environment and imaging conditions to ensure accurate results.
- Another critical precaution during imaging is to avoid photobleaching or photon pile-up, especially when adjusting the Z height to find the plane. Proper adjustment of laser power is essential to minimize these effects and ensure precise measurements. In addition, we discovered that cell incubation for at least 24 hours post-transfection of the reporter plasmid is necessary to capture robust differences between samples transfected with plasmids producing the RNA “A tags” versus RNA “D tags”.
- Furthermore, a pro-tip we learned is the significance of thoroughly washing off the glass beads used for bead loading the probe into cells. Imaging becomes more complex and challenging if any glass beads are left on the surface. We recommend multiple washes with PBS and tilting the plate at an angle to facilitate flow off and ensure optimal imaging conditions.
This study evaluates two riboswitches (A tag and D tag) that both bind Cbl tightly and demonstrate clear fluorescence lifetime differences (Figure 1g).2,13,14 Many more sequences of riboswitches from phylogeny are predicted to lead to a range of lifetimes. These efforts are currently being explored in our lab.15
In conclusion, our system offers several complementary advantages over other RNA labeling and visualization technologies. By utilizing a single laser and channel of detection and leveraging the versatility of riboswitch variants, we can control the lifetime of the RNA tag and use lifetime as the central mode of detection. With a wide range of available tags, we can label numerous RNAs and easily visualize them as our library of tags expands. In contrast, other technologies typically require multiple channels of detection and lasers, limiting expansion and multiplexing capabilities. Riboglow-FLIM analysis is a RNA tracking technology that offers several advantages. Its high specificity, ability to multiplex RNA imaging, use of only one laser, and channel of detection (as only one probe is being used) make it a valuable tool for studying dynamic RNA processes in living cells. Riboglow-FLIM has the potential to greatly advance our understanding of RNA dynamics and its role in various biological processes through system multiplexing.
Citations:
(1) Braselmann, E.; Rathbun, C.; Richards, E. M.; Palmer, A. E. Illuminating RNA Biology: Tools for Imaging RNA in Live Mammalian Cells. Cell Chemical Biology 2020, 27 (8), 891–903. https://doi.org/10.1016/j.chembiol.2020.06.010.
(2) Sarfraz, N.; Moscoso, E.; Oertel, T.; Lee, H. J.; Ranjit, S.; Braselmann, E. Visualizing Orthogonal RNAs Simultaneously in Live Mammalian Cells by Fluorescence Lifetime Imaging Microscopy (FLIM). Nature Communications 2023, 14 (1), 867. https://doi.org/10.1038/s41467-023-36531-y.
(3) Sunbul, M.; Lackner, J.; Martin, A.; Englert, D.; Hacene, B.; Grün, F.; Nienhaus, K.; Nienhaus, G. U.; Jäschke, A. Super-Resolution RNA Imaging Using a Rhodamine-Binding Aptamer with Fast Exchange Kinetics. Nature Biotechnology 2021, 39 (6), 686–690. https://doi.org/10.1038/s41587-020-00794-3.
(4) Paige, J. S.; Wu, K. Y.; Jaffrey, S. R. RNA Mimics of Green Fluorescent Protein. Science (New York, N.Y.) 2011, 333 (6042), 642—646. https://doi.org/10.1126/science.1207339.
(5) Filonov, G. S.; Moon, J. D.; Svensen, N.; Jaffrey, S. R. Broccoli: Rapid Selection of an RNA Mimic of Green Fluorescent Protein by Fluorescence-Based Selection and Directed Evolution. J. Am. Chem. Soc. 2014, 136 (46), 16299–16308. https://doi.org/10.1021/ja508478x.
(6) Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S. M.; Singer, R. H.; Long, R. M. Localization of ASH1 MRNA Particles in Living Yeast. Molecular Cell 1998, 2 (4), 437–445. https://doi.org/10.1016/S1097-2765(00)80143-4.
(7) Dolgosheina, E. V.; Jeng, S. C. Y.; Panchapakesan, S. S. S.; Cojocaru, R.; Chen, P. S. K.; Wilson, P. D.; Hawkins, N.; Wiggins, P. A.; Unrau, P. J. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chemical Biology 2014, 9 (10), 2412–2420. https://doi.org/10.1021/cb500499x.
(8) Chen, X.; Zhang, D.; Su, N.; Bao, B.; Xie, X.; Zuo, F.; Yang, L.; Wang, H.; Jiang, L.; Lin, Q.; Fang, M.; Li, N.; Hua, X.; Chen, Z.; Bao, C.; Xu, J.; Du, W.; Zhang, L.; Zhao, Y.; Zhu, L.; Loscalzo, J.; Yang, Y. Visualizing RNA Dynamics in Live Cells with Bright and Stable Fluorescent RNAs. Nature Biotechnology 2019, 37 (11), 1287–1293. https://doi.org/10.1038/s41587-019-0249-1.
(9) Bouhedda, F.; Fam, K. T.; Collot, M.; Autour, A.; Marzi, S.; Klymchenko, A.; Ryckelynck, M. A Dimerization-Based Fluorogenic Dye-Aptamer Module for RNA Imaging in Live Cells. Nature Chemical Biology 2020, 16 (1), 69–76. https://doi.org/10.1038/s41589-019-0381-8.
(10) Sunbul, M.; Jäschke, A. Contact-Mediated Quenching for RNA Imaging in Bacteria with a Fluorophore-Binding Aptamer. Angewandte Chemie (International ed. in English) 2013, 52. https://doi.org/10.1002/anie.201306622.
(11) Arora, A.; Sunbul, M.; Jäschke, A. Dual-Colour Imaging of RNAs Using Quencher- and Fluorophore-Binding Aptamers. Nucleic Acids Research 2015, 43 (21), e144–e144. https://doi.org/10.1093/nar/gkv718.
(12) Braselmann, E.; Palmer, A. E. A Multicolor Riboswitch-Based Platform for Imaging of RNA in Live Mammalian Cells. Methods Enzymol 2020, 641, 343–372. https://doi.org/10.1016/bs.mie.2020.03.004.
(13) Braselmann, E.; Wierzba, A. J.; Polaski, J. T.; Chromiński, M.; Holmes, Z. E.; Hung, S.-T.; Batan, D.; Wheeler, J. R.; Parker, R.; Jimenez, R.; Gryko, D.; Batey, R. T.; Palmer, A. E. A Multicolor Riboswitch-Based Platform for Imaging of RNA in Live Mammalian Cells. Nature Chemical Biology 2018, 14 (10), 964–971. https://doi.org/10.1038/s41589-018-0103-7.
(14) Polaski, J. T.; Webster, S. M.; Johnson, J. E.; Batey, R. T. Cobalamin Riboswitches Exhibit a Broad Range of Ability to Discriminate between Methylcobalamin and Adenosylcobalamin. Journal of Biological Chemistry 2017, 292 (28), 11650–11658. https://doi.org/10.1074/jbc.M117.787176.
(15) Rfam 14.7 (4069 Families), 2021. http://rfam.xfam.org/family/RF00174#tabview=tab1.


 (No Ratings Yet)
(No Ratings Yet)