Microscopy preprints – new tools and techniques in imaging and bioimage analysis
Posted by FocalPlane, on 3 May 2023
Here is a curated selection of preprints published recently. This post is a bumper edition, featuring preprints on bioimage analysis and new tools and techniques in imaging.
Bioimage analysis
Slitflow: a Python framework for single-molecule dynamics and localization analysis
Yuma Ito, Masanori Hirose, Makio Tokunaga
StereoCell enables highly accurate single-cell segmentation for spatial transcriptomics
Mei Li, Huanlin Liu, Min Li, Shuangsang Fang, Qiang Kang, Jiajun Zhang, Fei Teng, Dan Wang, Weixuan Cen, Zepeng Li, Ning Feng, Jing Guo, Qiqi He, Leying Wang, Tiantong Zheng, Shengkang Li, Yinqi Bai, Min Xie, Yong Bai, Sha Liao, Ao Chen, Susanne Brix, Xun Xu, Yong Zhang, Yuxiang Li
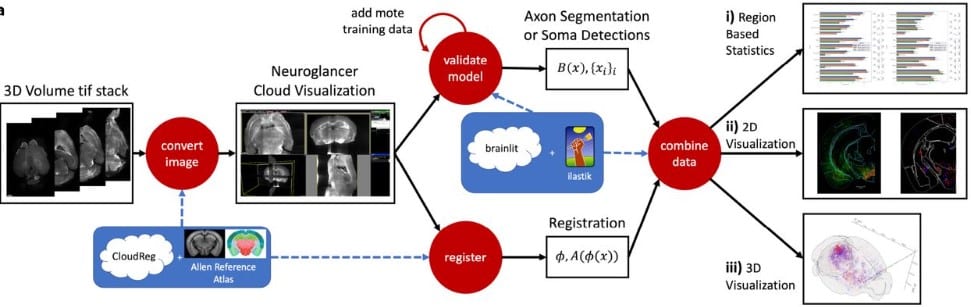
BrainLine: An Open Pipeline for Connectivity Analysis of Heterogeneous Whole-Brain Fluorescence Volumes
Thomas L. Athey, Matthew A. Wright, Marija Pavlovic, Vikram Chandrashekhar, Karl Deisseroth, Michael I. Miller, Joshua T. Vogelstein
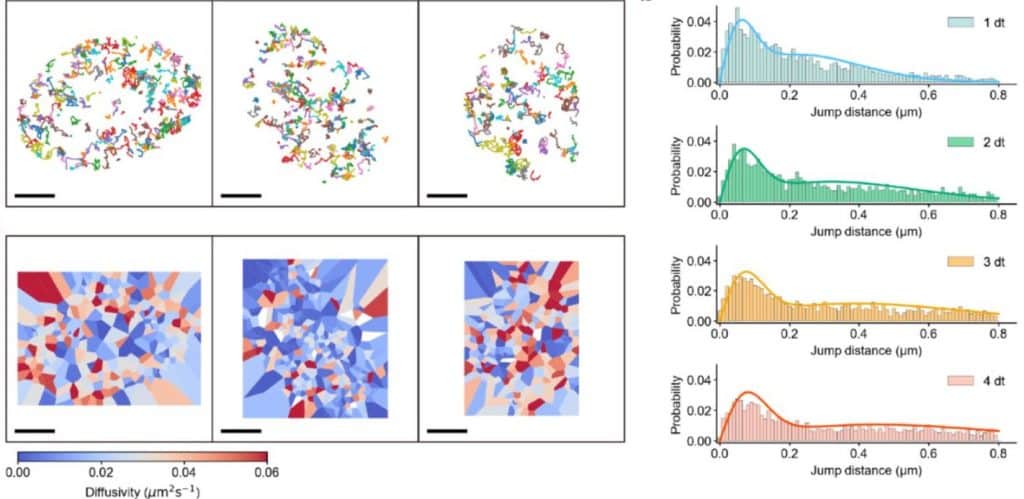
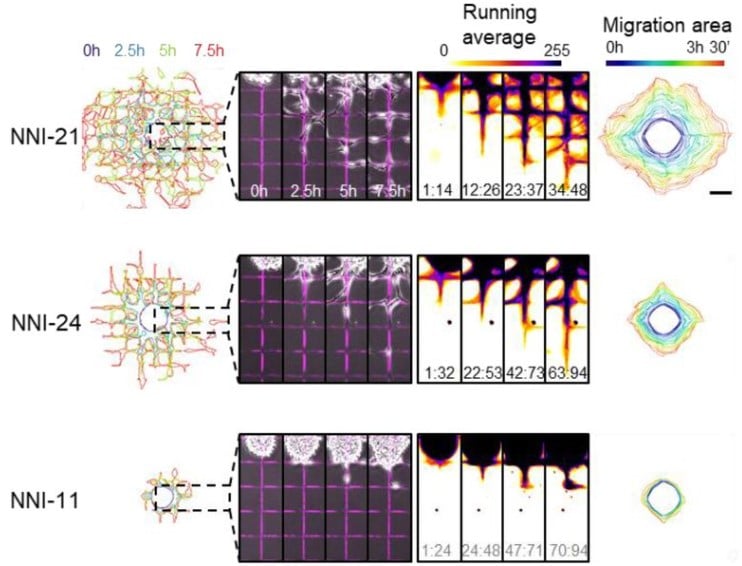
SP2G: an imaging and analysis pipeline revealing the inter and intra-patient migratory diversity of glioblastoma
Michele Crestani, Nikolaos Kakogiannos, Fabio Iannelli, Tania Dini, Claudio Maderna, Monica Giannotta, Giuliana Pelicci, Paolo Maiuri, Pascale Monzo, Nils C. Gauthier
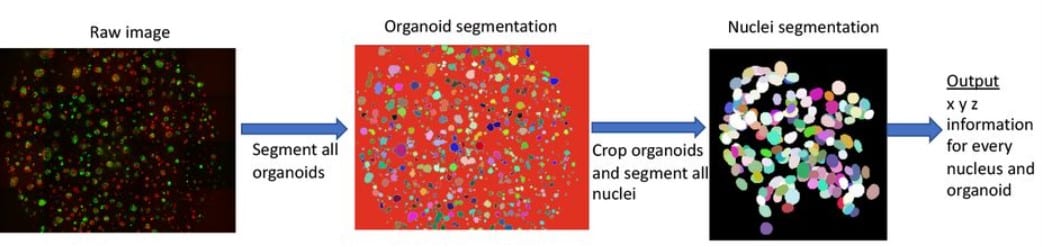
Cellos: High-throughput deconvolution of 3D organoid dynamics at cellular resolution for cancer pharmacology
Patience Mukashyaka, Pooja Kumar, David J. Mellert, Shadae Nicholas, Javad Noorbakhsh, Mattia Brugiolo, Olga Anczukow, Edison T. Liu, Jeffrey H. Chuang
Logic-based mechanistic machine learning on high-content images reveals how drugs differentially regulate cardiac fibroblasts
Anders R. Nelson, Steven L. Christiansen, Kristen M. Naegle, Jeffrey J. Saucerman
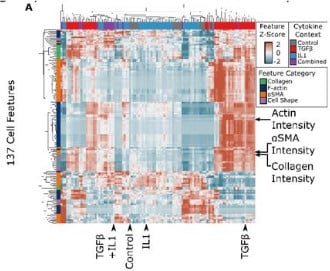
Figure extracted from Nelson, et al.
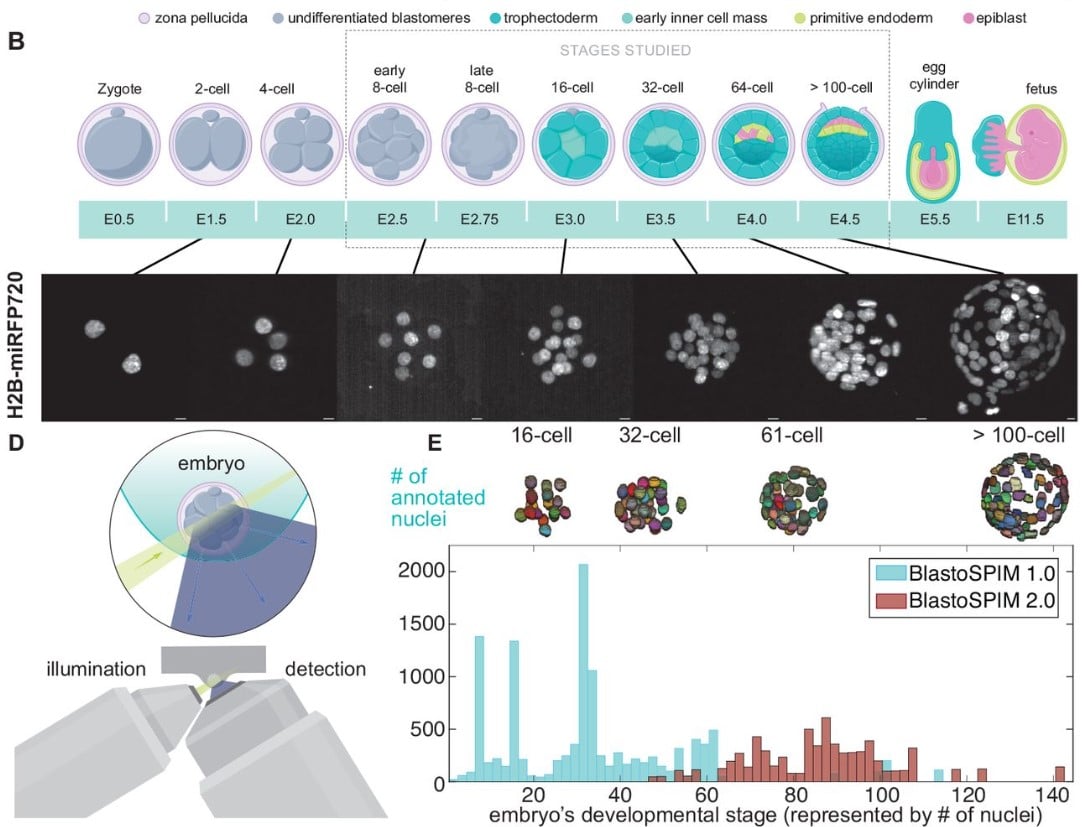
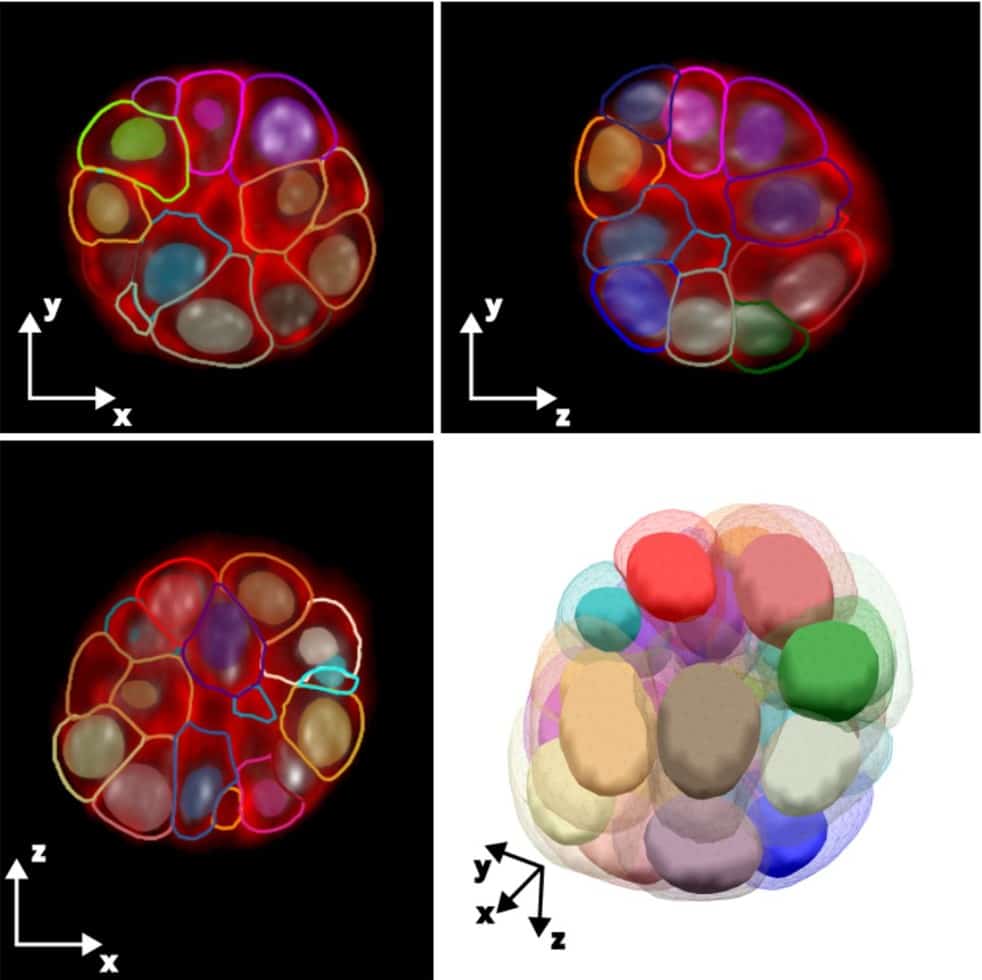
A novel ground truth dataset enables robust 3D nuclear instance segmentation in early mouse embryos
Hayden Nunley, Binglun Shao, Prateek Grover, Jaspreet Singh, Bradley Joyce, Rebecca Kim-Yip, Abraham Kohrman, Aaron Watters, Zsombor Gal, Alison Kickuth, Madeleine Chalifoux, Stanislav Shvartsman, Eszter Posfai, Lisa M. Brown
Noise-robust, physical microscopic deconvolution algorithm enabled by multi-resolution analysis regularization
Yiwei Hou, Wenyi Wang, Yunzhe Fu, Xichuan Ge, Meiqi Li, Peng Xi
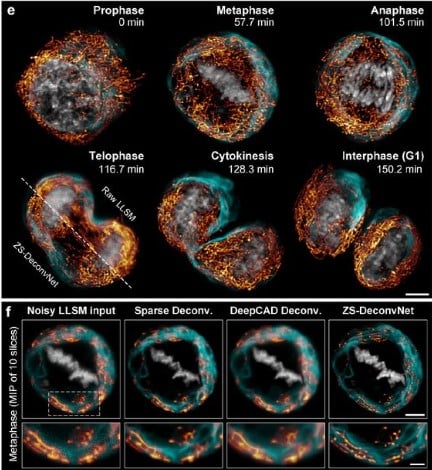
Zero-shot learning enables instant denoising and super-resolution in optical fluorescence microscopy
Chang Qiao, Yunmin Zeng, Quan Meng, Xingye Chen, Haoyu Chen, Tao Jiang, Rongfei Wei, Jiabao Guo, Wenfeng Fu, Huaide Lu, Di Li, Yuwang Wang, Hui Qiao, Jiamin Wu, Dong Li, Qionghai Dai
A fast algorithm for 3D volume reconstruction from light field microscopy datasets
Jonathan M. Taylor
Active mesh and neural network pipeline for cell aggregate segmentation
Matthew B. Smith, Hugh Sparks, Jorge Almagro, Agathe Chaigne, Axel Behrens, Chris Dunsby, Guillaume Salbreux
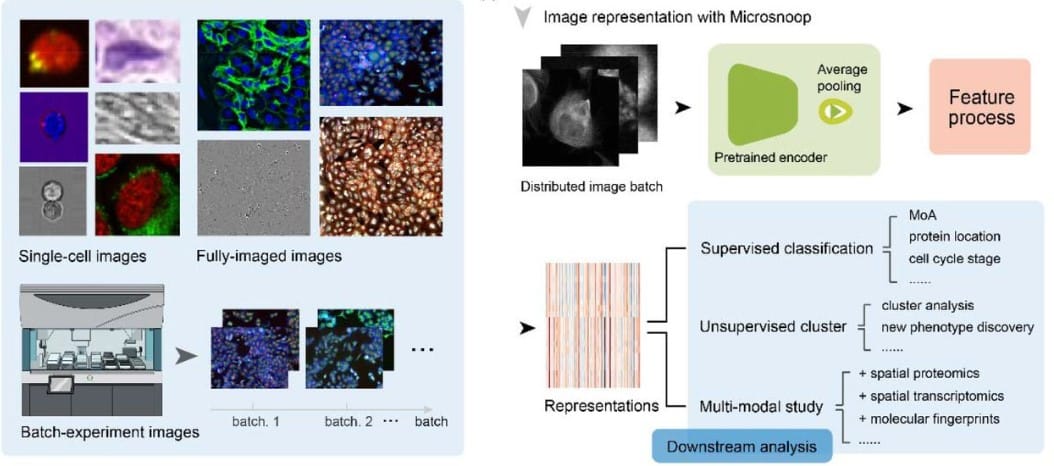
Microsnoop: a generalist tool for the unbiased representation of heterogeneous microscopy images
Dejin Xun, Rui Wang, Xingcai Zhang, Yi Wang
Dynamic superresolution by Bayesian nonparametric image processing
Ioannis Sgouralis, Ameya P. Jalihal, Lance W.Q. Xu, Nils G. Walter, Steve Pressé
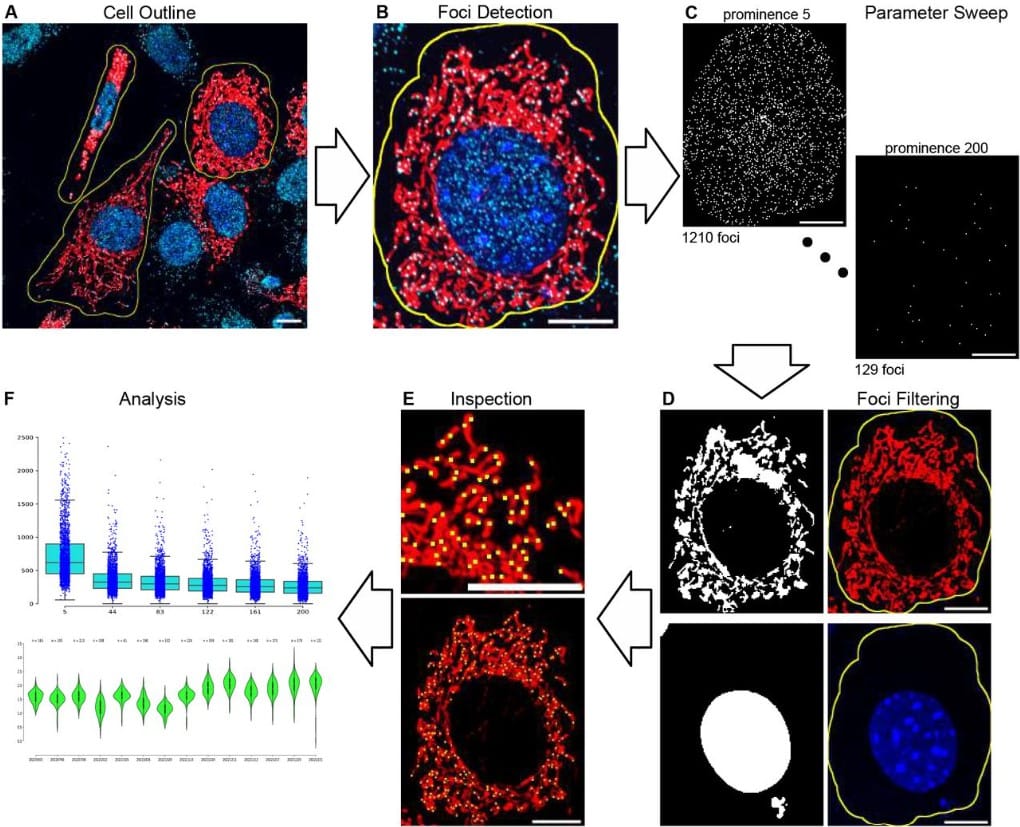
mtFociCounter for automated single-cell mitochondrial nucleoid quantification and reproducible foci analysis
Timo Rey, Luis Carlos Tábara, Julien Prudent, Michal Minczuk
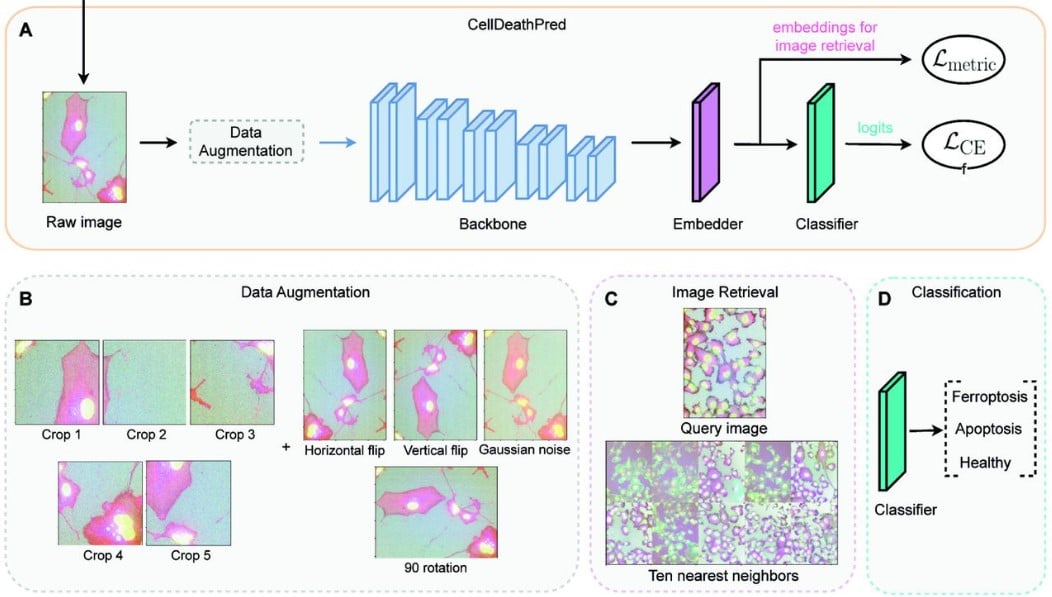
CellDeathPred: A Deep Learning framework for Ferroptosis and Apoptosis prediction based on cell painting
Kenji Schorpp, Alaa Bessadok, Aidin Biibosunov, Ina Rothenaigner, Stefanie Strasser, Tingying Peng, Kamyar Hadian
Self-supervised denoising for structured illumination microscopy enables long-term super-resolution live-cell imaging
Xingye Chen, Chang Qiao, Tao Jiang, Jiahao Liu, Quan Meng, Yunmin Zeng, Haoyu Chen, Yuanlong Zhang, Xinyang Li, Guoxun Zhang, Yixin Li, Hui Qiao, Jiamin Wu, Shan Tan, Dong Li, Qionghai Dai
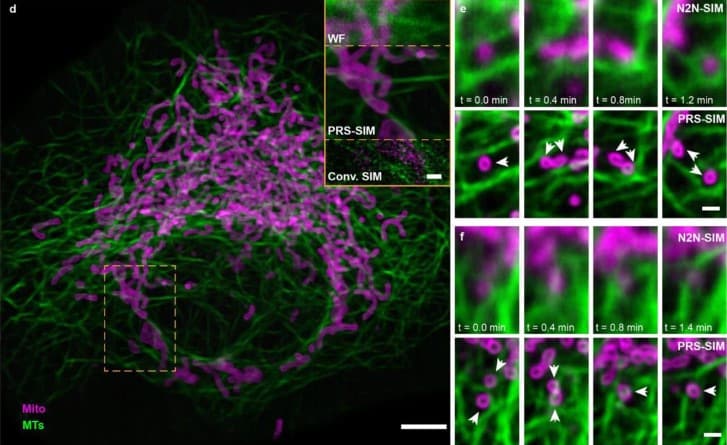
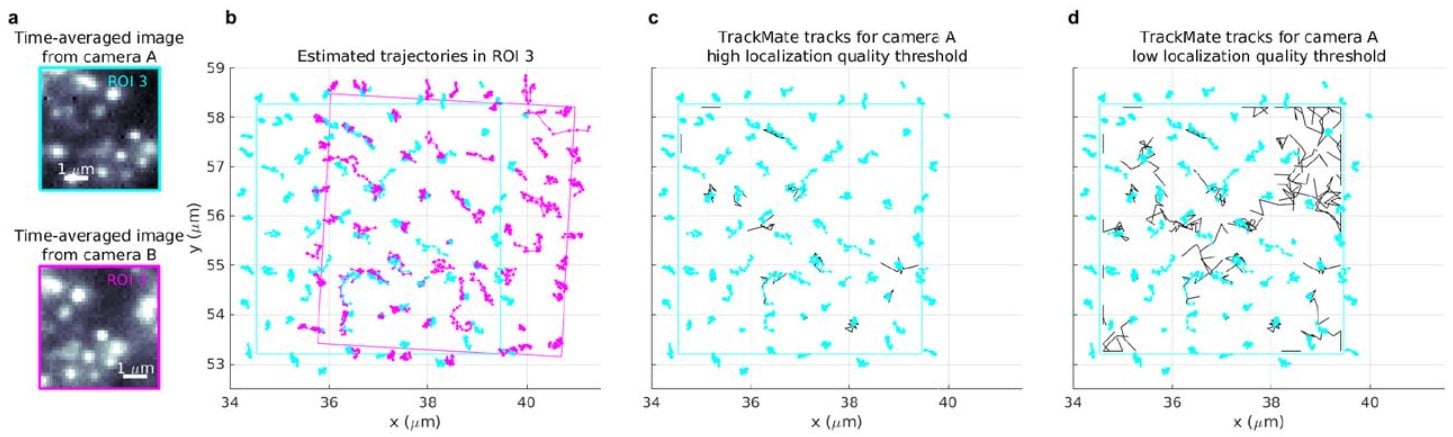
BNP-Track: A framework for multi-particle superresolved tracking
Lance W.Q. Xu, Ioannis Sgouralis, Zeliha Kilic, Steve Pressé
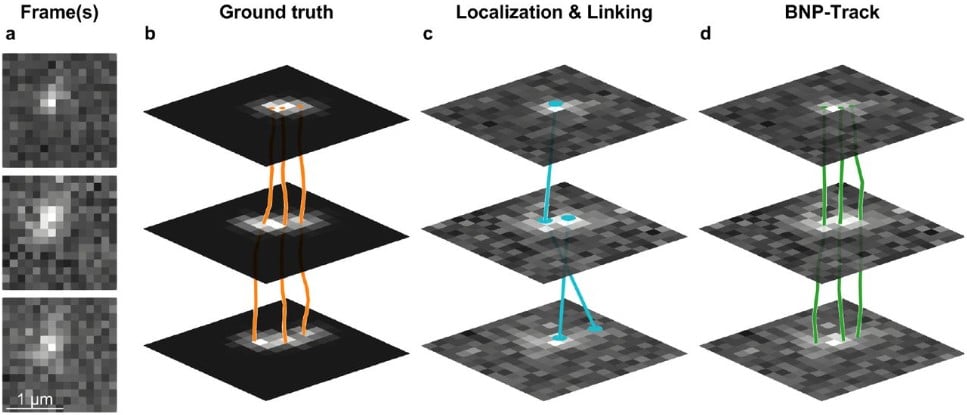
CebraEM: A practical workflow to segment cellular organelles in volume SEM datasets using a transferable CNN-based membrane prediction
Julian Hennies, José Miguel Serra Lleti, Constantin Pape, Sultan Bekbayev, Viktoriia Gross, Anna Kreshuk, Yannick Schwab
FLUTE: a Python GUI for interactive phasor analysis of FLIM data
Dale Gottlieb, Bahar Asadipour, Thi Phuong Lien Ung, Chiara Stringari
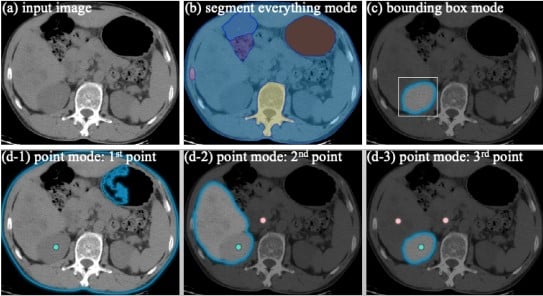
Segment Anything in Medical Images
Jun Ma, Bo Wang
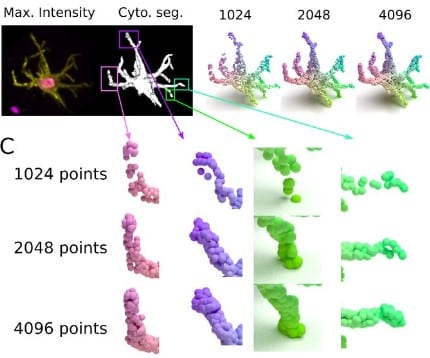
3D single-cell shape analysis using geometric deep learning
Matt De Vries, Lucas Dent, Nathan Curry, Leo Rowe-Brown, Vicky Bousgouni, Adam Tyson, Christopher Dunsby, Chris Bakal
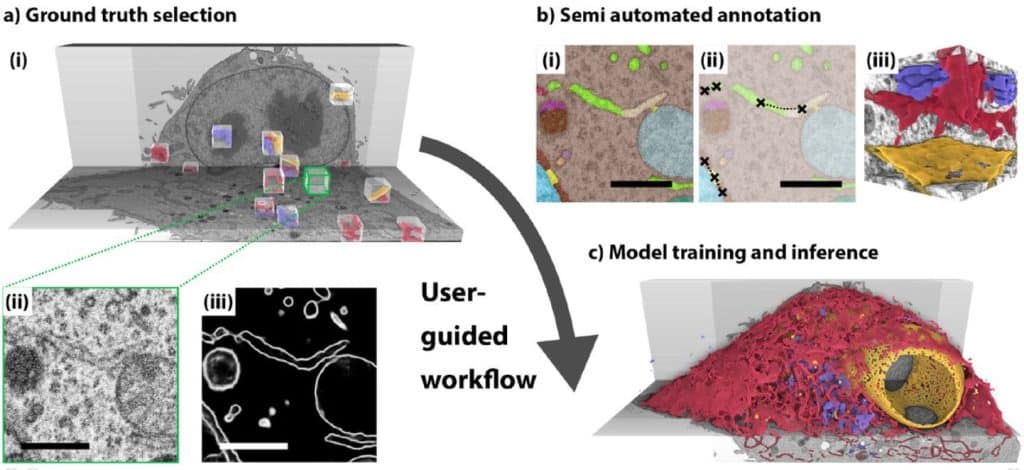
Organelle-specific segmentation, spatial analysis, and visualization of volume electron microscopy datasets
Andreas Müller, Deborah Schmidt, Lucas Rieckert, Michele Solimena, Martin Weigert
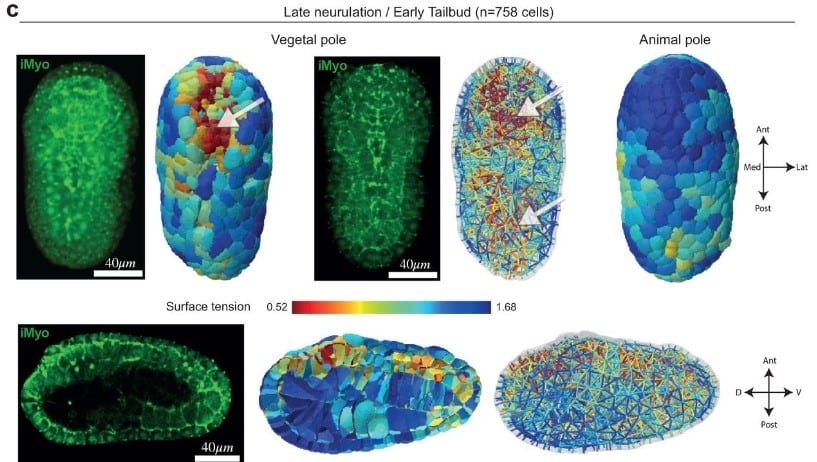
Embryo mechanics cartography: inference of 3D force atlases from fluorescence microscopy
Sacha Ichbiah, Fabrice Delbary, Alex McDougall, Rémi Dumollard, Hervé Turlier
Fluorescence Microscopy: a statistics-optics perspective
Mohamadreza Fazel, Kristin S. Grussmayer, Boris Ferdman, Aleksandra Radenovic, Yoav Shechtman, Jörg Enderlein, Steve Pressé
New tools and techniques
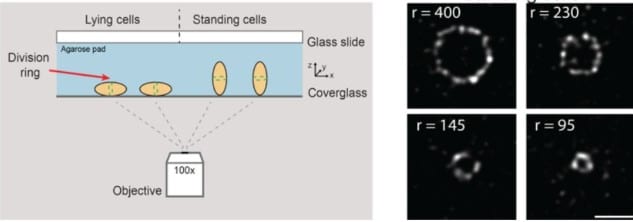
Application of nanotags and nanobodies for live cell single-molecule imaging of the Z-ring in Escherichia coli
Emma Westlund, Axel Bergenstråle, Alaska Pokhrel, Helena Chan, Ulf Skoglund, Daniel O. Daley, Bill Söderström
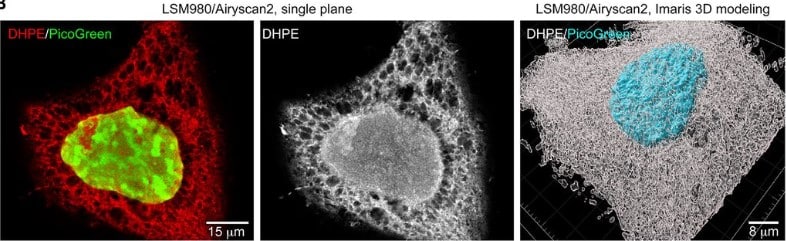
Expansion microscopy with trypsin digestion and tyramide signal amplification (TT-ExM) for protein and lipid staining
Ueh-Ting Tim Wang, Xuejiao Tian, Yae-Huei Liou, Sue-Ping Lee, Chieh-Han Lu, Peilin Chen, Bi-Chang Cheb
Thermogenetic control of Ca2+ levels in cells and tissues
Yulia G. Ermakova, Rainer Waadt, Mehmet S. Ozturk, Matvey Roshchin, Aleksandr A. Lanin, Artem Chebotarev, Matvey Pochechuev, Valeriy Pak, Ilya Kelmanson, Daria Smolyarova, Kaya Keutler, Alexander M. Matyushenko, Christian Tischer, Pavel M. Balaban, Evgeniy S. Nikitin, Karin Schumacher, Aleksei M. Zheltikov, Robert Prevedel, Carsten Schultz, Vsevolod V. Belousov
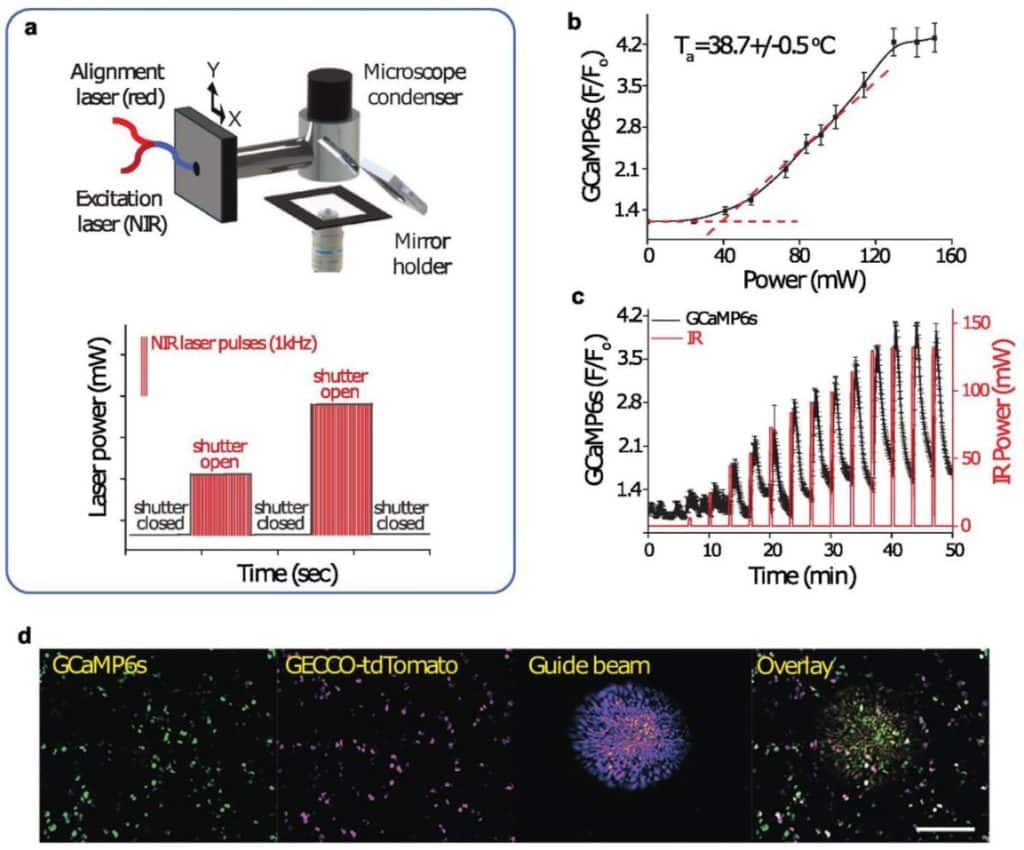
Optics-aware super-resolution light-field microscopy for long-term volumetric imaging of dynamic intracellular processes at millisecond timescales
Lanxin Zhu, Jiahao Sun, Chengqiang Yi, Meng Zhang, Guangda Niu, Jiang Tang, Yuhui Zhang, Dongyu Li, Peng Fei
Visualizing proteins by expansion microscopy
Ali H. Shaib, Abed Alrahman Chouaib, Rajdeep Chowdhury, Daniel Mihaylov, Chi Zhang, Vanessa Imani, Svilen Veselinov Georgiev, Nikolaos Mougios, Mehar Monga, Sofiia Reshetniak, Tiago Mimoso, Han Chen, Parisa Fatehbasharzad, Dagmar Crzan, Kim-Ann Saal, Nadia Alawar, Janna Eilts, Jinyoung Kang, Luis Alvarez, Claudia Trenkwalder, Brit Mollenhauer, Tiago F. Outeiro, Sarah Köster, Julia Preobraschenski, Ute Becherer, Tobias Moser, Edward S. Boyden, A Radu Aricescu, Markus Sauer, Felipe Opazo, Silvio O. Rizzoli
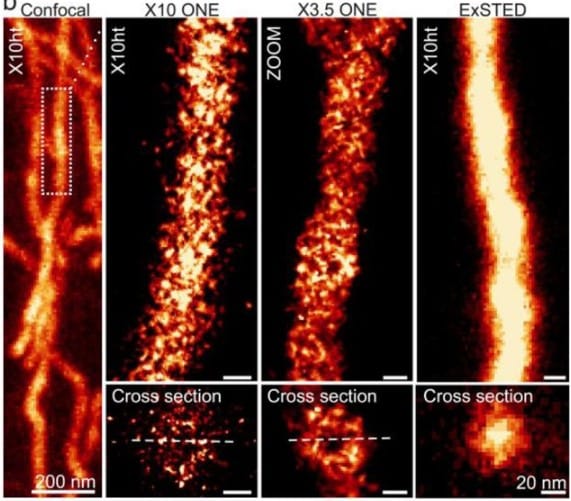
Smart Lattice Light Sheet Microscopy for imaging rare and complex cellular events
Yu Shi, Jimmy S. Tabet, Daniel E. Milkie, Timothy A. Daugird, Chelsea Q. Yang, Andrea Giovannucci, Wesley R. Legant
Hyperspectral Oblique Plane Microscopy
Ke Guo, Konstantinos Kalyviotis, Periklis Pantazis, Christopher J Rowlands
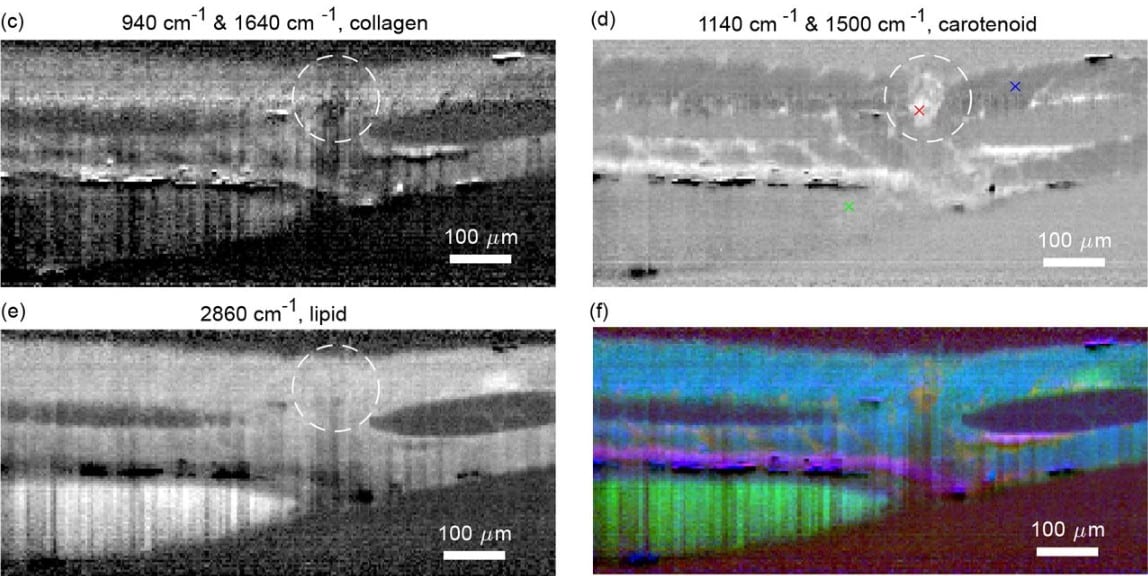
The TriScan: fast and sensitive 3D confocal fluorescence imaging using a simple optical design
Robin Van den Eynde, Wim Vandenberg, Peter Dedecker
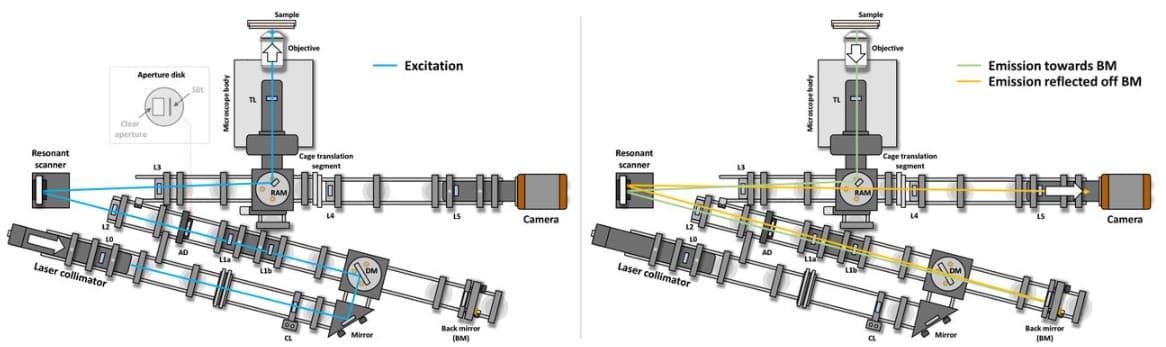
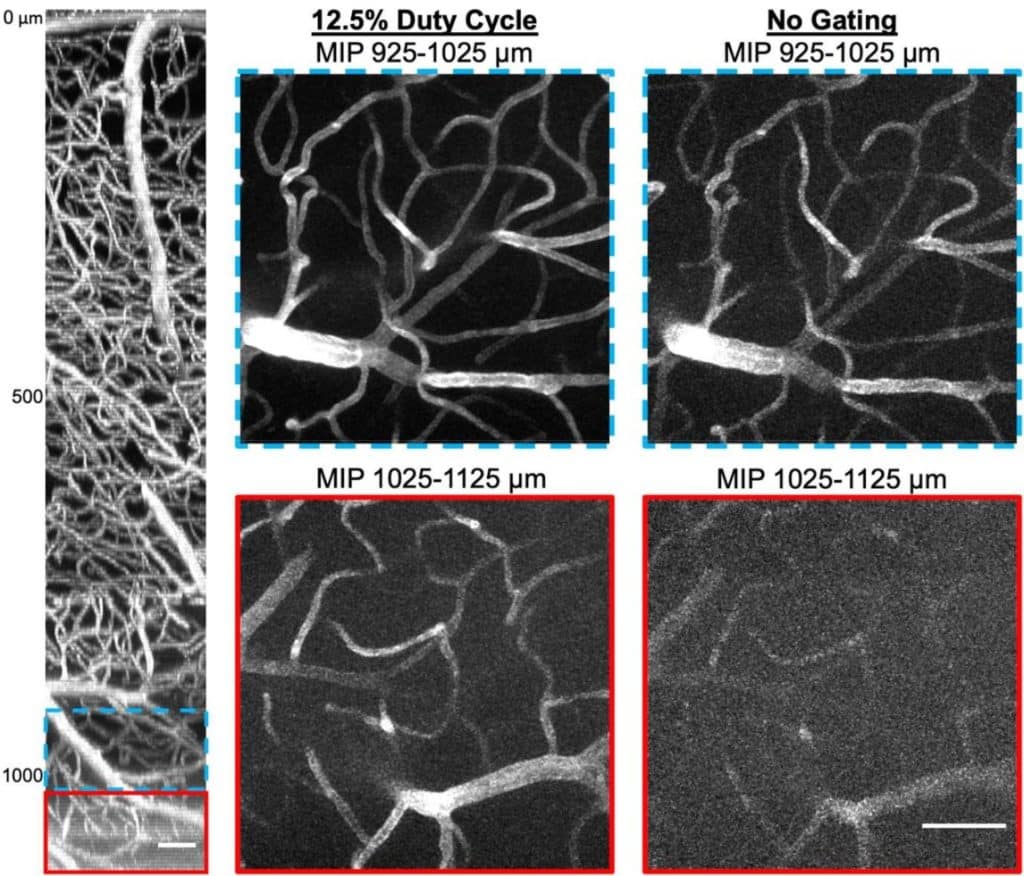
Pulse train gating to improve signal generation for in vivo two-photon fluorescence microscopy
Shaun A. Engelmann, Alankrit Tomar, Aaron L. Woods, Andrew K. Dunn
Validating a low-cost, open-source, locally manufactured workstation and computational pipeline for automated histopathology evaluation using deep learning
Divya Choudhury, James Dolezal, Emma Dyer, Sara Kochanny, Siddi Ramesh, Frederick M. Howard, Jayson R. Margalus, Amelia Schroeder, Jefree Schulte, Marina C. Garassino, Jakob N. Kather, Alexander T. Pearson
Video-rate three-photon imaging in deep Drosophila brain based on a single Cr:forsterite oscillator
Lu-Ting Chou, Shao-Hsuan Wu, Hao-Hsuan Hung, Je-Chi Jang, Chung-Ming Chen, Ting-Chen Chang, Wei-Zhong Lin, Li-An Chu, Chi-Kuang Sun, Franz X. Kärtner, Anatoly A. Ivanov, Shi-Wei Chu, Shih-Hsuan Chia
Tunable photoinitiated hydrogel microspheres for direct quantification of cell-generated forces in complex three-dimensional environments
Antoni Garcia-Herreros, Yi-Ting Yeh, Yunpeng Tu, Adithan Kandasamy, Juan C. del Alamo, Ernesto Criado-Hidalgo
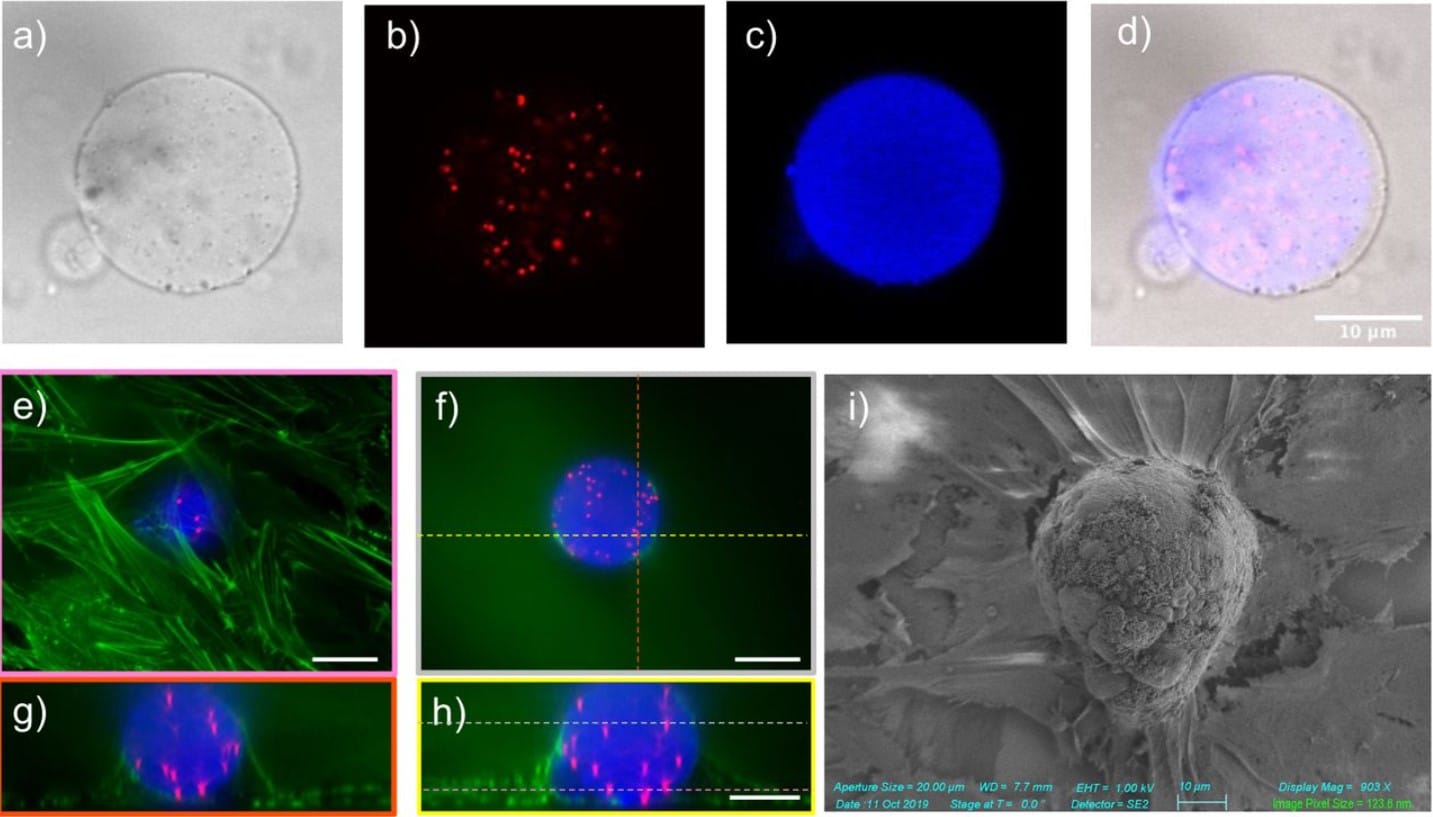
Large tissue archiving solution for multiplexed labeling and super-resolution imaging
Ya-Hui Lin, Li-Wen Wang, Yen-Hui Chen, Yi-Chieh Chan, Shang-Hsiu Hu, Sheng-Yan Wu, Chi-Shiun Chiang, Guan-Jie Huang, Shang-Da Yang, Shi-Wei Chu, Kuo-Chuan Wang, Chin-Hsien Lin, Pei-Hsin Huang, Hwai-Jong Cheng, Bi-Chang Chen, Li-An Chu
On-grid purification of electron microscopy samples via a 3D-printed flow-cell
Kailash Ramlaul, Ziyi Feng, Caoimhe Canavan, Natàlia de Martín Garrido, David Carreño, Michael Crone, Kirsten E. Jensen, Bing Li, Harry Barnet, David T. Riglar, David Miller, Paul S. Freemont, Christopher H. S. Aylett
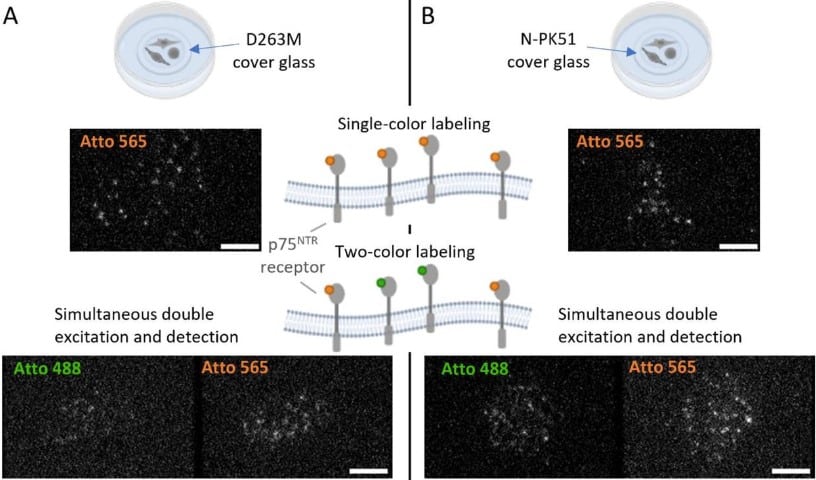
Optimized two-color single-molecule tracking of fast-diffusing membrane receptors
Chiara Schirripa Spagnolo, Aldo Moscardini, Rosy Amodeo, Fabio Beltram, Stefano Luin
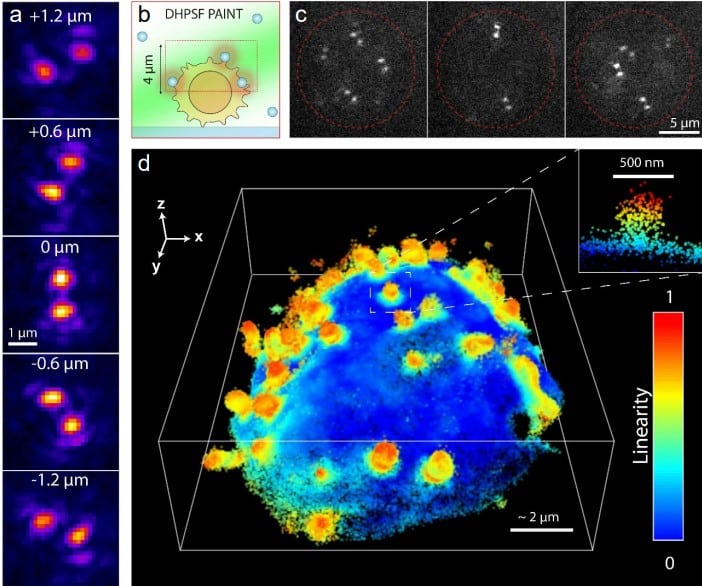
Additive manufacturing of solid diffractive optical elements via near index matching
Reut Kedem Orange, Nadav Opatovski, Dafei Xiao, Boris Ferdman, Onit Alalouf, Sushanta Kumar Pal, Ziyun Wang, Henrik von der Emde, Michael Weber, Steffen J. Sahl, Aleks Ponjavic, Ady Arie, Stefan W. Hell, Yoav Shechtman
RoPod, a customizable toolkit for non-invasive root imaging, reveals cell type-specific dynamics of plant autophagy
Marjorie Guichard, Sanjana Holla, Daša Wernerová, Guido Grossmann, Elena A. Minina

Quantitative determination of fluorescence labeling implemented in cell cultures
Chiara Schirripa Spagnolo, Aldo Moscardini, Rosy Amodeo, Fabio Beltram, Stefano Luin
Fluorescent Sensors for Imaging Interstitial Calcium
Ariel A. Valiente-Gabioud, Inés Garteizgogeascoa Suñer, Agata Idziak, Arne Fabritius, Julie Angibaud, Jérome Basquin, U. Valentin Nägerl, Sumeet Pal Singh, Oliver Griesbeck
Super-Resolved FRET and Co-Tracking in pMINFLUX
Fiona Cole, Jonas Zähringer, Johann Bohlen, Tim Schröder, Florian Steiner, Fernando D. Stefani, Philip Tinnefeld
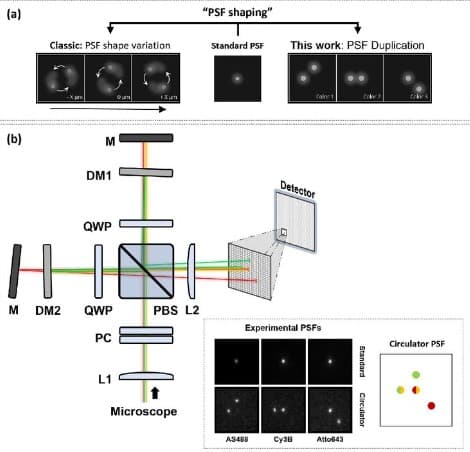
Simultaneous multicolor fluorescence imaging using duplication-based PSF engineering
Robin Van den Eynde, Fabian Hertel, Bartosz Krajnik, Siewert Hugelier, Alexander Auer, Thomas Schlichthaerle, Ralf Jungmann, Marcel Leutenegger, Wim Vandenberg, Peter Dedecker


 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)