All things unequal – live microscopy reveals asymmetric RNA localization in the early mammalian embryo
Posted by Jennifer Zenker, on 4 September 2023
Authors:
Jennifer Zenker1,*, Julian Heng2
1Australian Regenerative Medicine Institute (ARMI), Faculty of Medicine, Nursing and Health Sciences, Monash University, Clayton, Australia
2Remotely Consulting, Perth, Australia
*Corresponding author
Non-mammalian systems have been invaluable for us to decipher evolutionarily-conserved processes, orthologous molecules and cellular phenomena underlying early embryo development. One fundamental discovery from the work on non-mammalian systems has been that the distinct localization patterns for intracellular factors within specific cells of the developing embryo are essential to guide embryonic cell differentiation and cell fate determination. Until a decade ago, technical limitations of microscopes made studying mammalian embryos and their constituent cells a very difficult task. Adding to this challenge, methods to manipulate mammalian embryos and their constituent cells, as well as the ability for researchers to study distinct intracellular components such as RNAs, all while preserving embryos in laboratory conditions to watch them in action, were altogether lacking. It thus speaks to the remarkable progress collectively made by researchers in this field over the last ten years, including work from groups from the UK, USA, France, Canada, Australia and beyond, to have successfully adapted microscopy, ex vivo cell culture and genetic labelling techniques to ethically study living embryos from multiple mammalian species1-5. As it turns out, recent studies with mouse early-stage, preimplantation embryos have found that both the subtype of RNA and their localization within cells of the mouse embryo, have a lot to do with how each cell behaves as the embryo matures6,7.
Setting the stage – microscopy for imaging living mouse preimplantation embryos
Studying preimplantation embryos from mammals requires tools to visualize these small but opaque clumps of cells, as well as the means to study them ex vivo. Such technical challenges are well addressed by using imaging platforms such as the Confocal ZEISS LSM780 equipped with environmental chambers (Fig. 1A) and Avalanche Photodiodes (APDs)5,8, as well as the newest ZEISS Lattice Light Sheet 77. Rather than acquiring static, two-dimensional images of preserved specimens through conventional microscopy, images at high-resolution of living mouse embryos are continuously recorded in real-time, at predefined intervals, typically spanning hours to days, and across the depth of each specimen (Fig. 1B).
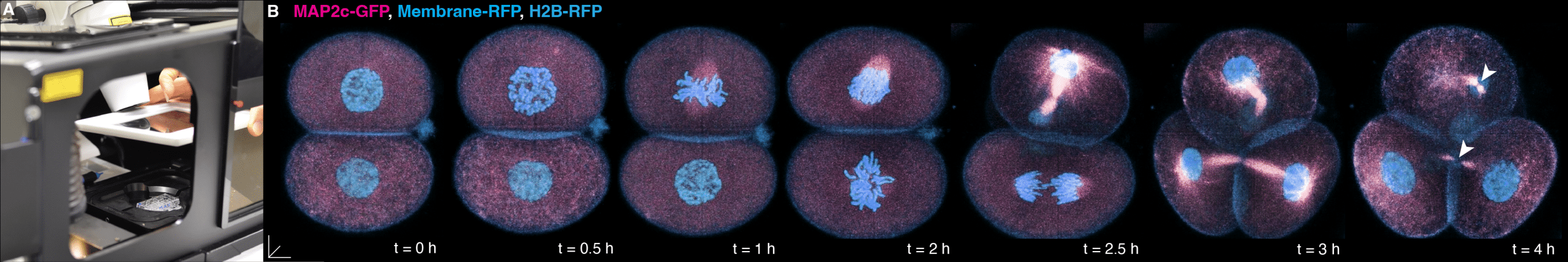
(A) Close-up photo of the ZEISS Confocal LSM 780 microscope during the mounting of the imaging chamber into the environmental chamber. (B) 4D imaging (x, y, z and time) as a living preimplantation mouse embryo divides from 2- to 4-cell stage, microinjected with fluorescently-tagged RNA constructs to visualise the microtubule cytoskeleton (MAP2c-GFP; pink), Membrane (Membrane-RFP; blue) and chromatin via a Histone 2B label (H2B-RFP; blue) (right, time-lapse panels). White arrowheads at t = 4h highlight the “interphase bridge”, serving as a non-centrosomal microtubule organising centre in the cells of the preimplantation embryo. Temporal resolution, every 15 min; Scale bars, 10 mm.
Maintaining the viability of the embryo during live imaging is of course a critical factor. Any mishandling or stress during the procedure may adversely affect the embryo and skew the results collected from these. To simulate, as much as possible, the environment inside the womb for these embryos, stable imaging conditions protected from intense (day)light must be maintained, while ensuring appropriate temperature, humidity, pH and levels of dissolved gases, such as oxygen and carbon dioxide, in culture media. We have found that even tiny fluctuations, of less than 1°C in temperature or of less than 1% gas mixture in the incubator, can compromise the development of the living embryo or the quality of imaging data acquired, or both. By applying such cutting-edge microscope technologies we observed, for the first time, that the microtubule cytoskeleton of cells within the mammalian preimplantation embryos is highly organised5, and that the asymmetric, intracellular organization of RNA subtypes (namely rRNAs, mRNAs and tRNAs) influences cell fate determination7, as explained further below.
Orchestrated from within – cell fate determination in the preimplantation embryo requires an organised microtubule cytoskeleton
Once we solved the challenge of maintaining preimplantation mouse embryos in an imaging chamber and how to make high-quality recordings of them in real-time, we wanted answers to the question that is asked by everyone in our field of work: how are the earliest cells of the mammalian embryo deciding how and what they want to become?
There were already some clues from research carried out some years earlier as to how to look for these decision-making factors. Studies from non-mammalian embryos had implicated the microtubule cytoskeleton within cells of early-stage embryos as critical components that influenced cell fate determination9, and so we took on such insights to begin our investigation. To our surprise, we discovered that, in cells of the mammalian (mouse) preimplantation embryo, the microtubule cytoskeleton was highly structured, which we characterized as a highly complex and asymmetric network of internal scaffolds5. This finding was remarkable at the time because, until our report, it was generally accepted that the microtubule cytoskeleton within cells of preimplantation mammalian embryos was largely disorganised10. We further hypothesised that this unprecedent form of microtubule organisation may also exist in cells from other mammalian preimplantation embryos, including humans11. Further to this, we discovered that the microtubule cytoskeleton was important to the intracellular processes for cell adhesion, fate determination and pluripotency5 (Fig. 2).

Following the creation of a microtubule roadmap (pink), the directional transport of cargo (represented by cars) can be investigated to clarify how the microtubule cytoskeleton is important to the regulation of cell fate and pluripotent cell states.
Light them up – labelling RNAs within preimplantation embryos to study their functions
That, however, was not where our studies ended. We knew that the internal cytoskeleton of cells can act as an ‘information highway’ to distribute molecular cargos to specific locations12, and so we asked: was the microtubule cytoskeleton the responsible agent for cell fate determination within the preimplantation embryo, or were they critical to facilitate the decision-making process? Given the wealth of information from studies of non-mammalian embryos showing that specific intracellular factors related to the microtubule cytoskeleton might be driving cell fate decisions9, we reasoned that some of this molecular cargo could in fact be the responsible agents. Indeed, the asymmetric localisation patterns for multiple RNA species have been found to be instructive for cell fate determination in the embryos of drosophila13, 14, xenopus15 and zebrafish embryos16, 17, and RNA is known to be trafficked throughout the cell of an embryo through its microtubule network9. After using conventional approaches including Fluorescence In Situ Hybridization (FISH) to label specific RNAs and confirm that RNA localisation, at least anecdotally, was asymmetric within cells of the preimplantation embryo, we then used innovative live dyes to, for the first time, image global RNAs (= various RNA types) in living mammalian embryos in real-time. We found that global RNA was indeed asymmetrically localised in a microtubule-dependent fashion when embryos were about to divide from the 16- to 32-cell stage (Fig. 3). To push the technology further, we also developed ways to image the entire pool of mRNA, rRNA18 and tRNA subtypes. To our delight, we found that different species of RNAs and their distinct location within cells influenced their cell fate decisions, whether they formed progenitors of the pluripotent inner cell mass forming all embryonic tissues or extraembryonic cells.
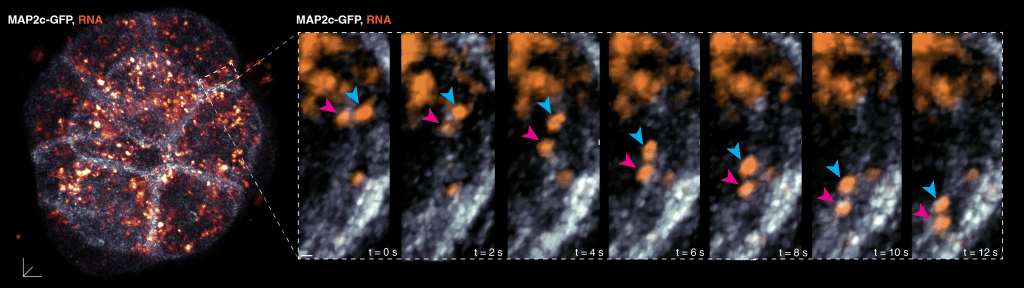
High-magnification, temporal real-time imaging in a region of interest (dashed boxes) of a live 16-cell stage mouse preimplantation embryo, labelled to visualise the microtubule cytoskeleton (MAP2c-GFP; white) and RNA (orange signals). 2 RNA foci tracked (magenta and blue arrowheads) showing unidirectional transport along a microtubule filament, leading to asymmetries in RNA localisation within the cell. Temporal resolution, every 1 second (s); Scale bar, 10 mm and 2 mm in insets.
We were thrilled by our results to image RNA in live mammalian embryos because we knew that, prior to our studies, RNA localisation was documented only anecdotally through the characterisation of selected gene transcripts using the FISH protocol on fixed samples. In addition to our findings, we also successfully adapted a molecular technique known as SunTag19,20 to visualise translation of gene-specific mRNAs within cells of the preimplantation embryo in real-time. This led us to discover that translation does not necessarily occur at areas of highest RNA abundance, but rather in areas where mRNAs, rRNAs and tRNAs are found together instead. This compartmentalisation of RNA subtypes and translation capability is then passed onto the next generation of cells as the embryo reaches 32-cell stage. Notably, cells that have decided to become extraembryonic tissue require a higher RNA translation capacity as compared to cells of the pluripotent inner cell mass, which are rather more undecided in their fates. Our findings thus bring into prominence the roles for different RNA sub-types in cell-fate decision-making, and broaden our understanding of the regulatory signalling hierarchies within early mammalian embryos.
Reflections of a research group studying living embryos in real-time
There are some peculiar aspects to working with preimplantation mouse embryos. For example, one needs to be in the lab at the right time which, to admit, can mean all the time, at least when an experiment is running. Working with living mouse embryos, it is also not possible to press the “stop button” when normal working hours are over. The period of interest for our work was predominantly during 16-cell stage interphase. With cell divisions occurring about every 12h, this period occurred in the early evening 2.5 days after mating, given that mice, which are nocturnally active, mate at night. For researchers, an extraordinary level of time management and commitment, and planned breaks, are all called for, for that incredible moment when sitting at the microscope to witness all that is happening within these embryos in real-time.
Another aspect to this work is the vast amount of data that is generated during live imaging, which needs to be processed and analysed effectively, typically as terabyte-sized files of scans that need to be aligned to reconstruct three-dimensional images of embryos to be able to document the intracellular events that are unfolding within. All this requires the use of advanced image processing techniques and a minimum level of experience of the researchers to perform time-consuming in-depth data analysis, and to extract meaningful information from the collected data. Also, live-imaging studies of embryos involves the use of mice, in this case, which raises ethical considerations. Researchers must follow strict ethical guidelines, ensure the welfare of the animals involved and use the minimal number of animals possible.
Studying preimplantation embryos to improve health
Innovations in this field of preimplantation mammalian embryogenesis are already leading us to consider new ways of defining the quality of embryos, in addition to conventional qualitative traits such as compaction, morphology by light microscopy and mitochondrial number. Live-imaging studies on embryonic development will also be crucial to clarify our understanding of the impact of disease-causing genetic variants that severely restrict lifespan, as well as how birth defects and congenital anomalies arise. Future studies to define pluripotency in cells of preimplantation embryos, as well as in human in vitro models21 could offer new possibilities in regenerative medicine research. Such efforts will lead to the development of improved diagnostics and therapeutic strategies that improve human health.
References:
1Rossant, J. Making the Mouse Blastocyst: Past, Presence, and Future. Current Topics in Developmental Biology 117, 275-288 (2016).
2Shabazi, MN. and Zernicka-Goetz, M. Deconstructing and reconstructing the mouse and human early embryo. Nature Cell Biology 20, 878-887 (2018).
3Dumortier, JG. et al. Hydraulic fracturing and active coarsening position the lumen of the mouse blastocyst. Science 365 (6452), 465-468 (2019).
4Vazquez-Diez, C. et al. Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. PNAS 113(3), 626-631 (2016).
5Zenker, J. et al. A microtubule-organizing center directing intracellular transport in the early mouse embryo. Science 357, 925–928 (2017).
6Skamagki, M. et al. Asymmetric localization of Cdx2 mRNA during the first cell-fate decision in early mouse development. Cell Rep. 3, 442–457 (2013).
7 Hawdon, A. et al., Apicobasal RNA asymmetries regulate cell fate in the early mouse embryo. Nature Communications 14(1): p. 2909 (2023).
8Zenker, J. et al. Expanding actin rings zipper the mouse embryo for blastocyst formation. Cell 173, 776–791.e717 (2018).
9Ephrussi, A. and Johnston, D. S. Seeing is believing: the bicoid morphogen gradient matures. Cell 116, 143–152 (2004).
10Howe K. and FitzHarris, G. A non-canonical mode of microtubule organization operates throughout preimplantation development in mouse. Cell Cycle (12), 1616-1624 (2013).
11Palacios Martinez S. et al. Beyond the centrosome: The mystery of microtubule organising centres across mammalian preimplantation embryos. Current Opinion in Cell Biology 77:102114 (2022).
12Barlan, K and Gelfand, VI, Microtubule-Based transport and the Distribution, Tethering, and Organization of Organelles. Cold Spring Harb Perspect Biol 9(5):a025817 (2017).
13Driever, W. & Nüsslein-Volhard, C. A gradient of bicoid protein in Drosophila embryos. Cell 54, 83–93 (1988).
14Dufourt, J. et al. Imaging translation dynamics in live embryos reveals spatial heterogeneities. Science 372, 840–844 (2021).
15Kloc, M. et al. Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Developmental biology 241, 79-93 (2002).
16Lunde, K. at al. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Current biology 14, 48-55 (2004).
17Abrams, EW. and Mullins, MC. Early zebrafish development: it’s in the maternal genes. Current opinion in genetics & development 19, 396-403 (2009).
18Cao, C. et al. Ribosomal RNA-selective light-up fluorescent probe for rapidly imaging the nucleolus in live cells. ACS Sens 4, 1409–1416 (2019).
19Tanenbaum, ME. et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
20Yan, X. et al. Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989 (2016).
21Liu X. et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021, 591:627–632 (2021).


 (2 votes, average: 1.00 out of 1)
(2 votes, average: 1.00 out of 1)