Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 6 October 2023
Here is a curated selection of preprints published recently. In this post, we focus specifically on new tools for bioimage analysis.
CartoCell, a high-content pipeline for 3D image analysis, unveils cell morphology patterns in epithelia
Jesús A. Andrés-San Román, Carmen Gordillo-Vázquez, Daniel Franco-Barranco, Laura Morato, Cecilia H. Fernández-Espartero, Gabriel Baonza, Antonio Tagua, Pablo Vicente-Munuera, Ana M. Palacios, María P. Gavilán, Fernando Martín-Belmonte, Valentina Annese, Pedro Gómez-Gálvez, Ignacio Arganda-Carreras, Luis M. Escudero
Phasor Identifier: A Cloud-based Analysis of Phasor-FLIM Data on Python Notebooks
Mario Bernardi, Francesco Cardarelli
Machine assisted annotation in neuroanatomy
Kui Qian, Beth Friedman, David Kleinfeld, Yoav Freund
Omega – Harnessing the Power of Large Language Models for Bioimage Analysis
Royer, Loic A.
Neural Network Informed Photon Filtering Reduces Artifacts in Fluorescence Correlation Spectroscopy Data
Alexander Seltmann, Pablo Carravilla, Katharina Reglinski, Christian Eggeling, Dominic Waithe
Resolution enhancement with deblurring by pixel reassignment (DPR)
Bingying Zhao, Jerome Mertz
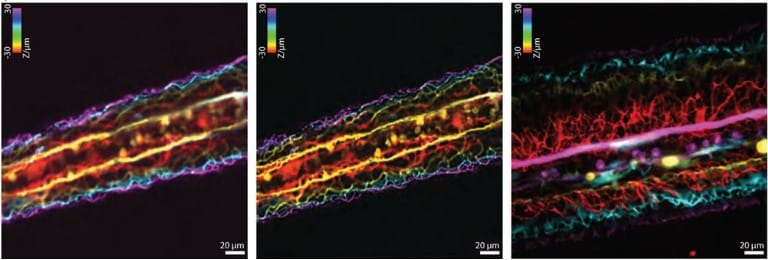
Open and reusable deep learning for pathology with WSInfer and QuPath
Jakub R. Kaczmarzyk, Alan O’Callaghan, Fiona Inglis, Tahsin Kurc, Rajarsi Gupta, Erich Bremer, Peter Bankhead, Joel H. Saltz
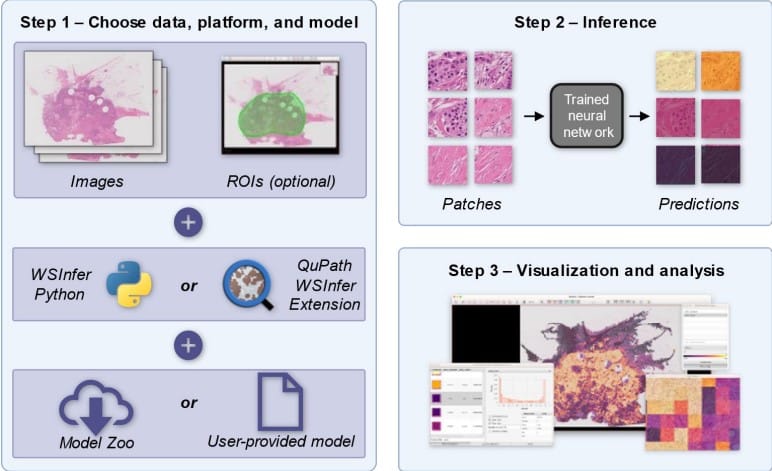
μSplit: efficient image decomposition for microscopy data
Ashesh, Alexander Krull, Moises Di Sante, Francesco Silvio Pasqualini, Florian Jug
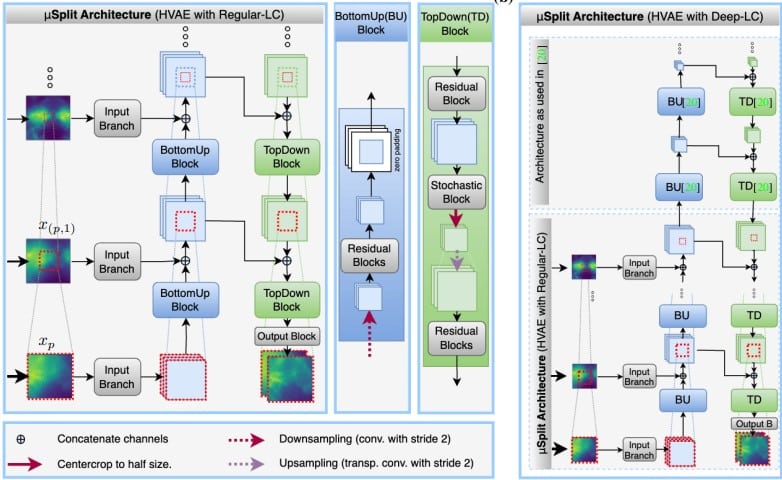
BigFUSE: Global Context-Aware Image Fusion in Dual-View Light-Sheet Fluorescence Microscopy with Image Formation Prior
Yu Liu, Gesine Muller, Nassir Navab, Carsten Marr, Jan Huisken, Tingying Peng
Spatial redundancy transformer for self-supervised fluorescence image denoising
Xinyang Li, Xiaowan Hu, Xingye Chen, Jiaqi Fan, Zhifeng Zhao, Jiamin Wu, Haoqian Wang, Qionghai Dai
High-fidelity Image Restoration of Large 3D Electron Microscopy Volume
Yuri Kreinin, Pat Gunn, Dmitri Chklovskii, Jingpeng Wu
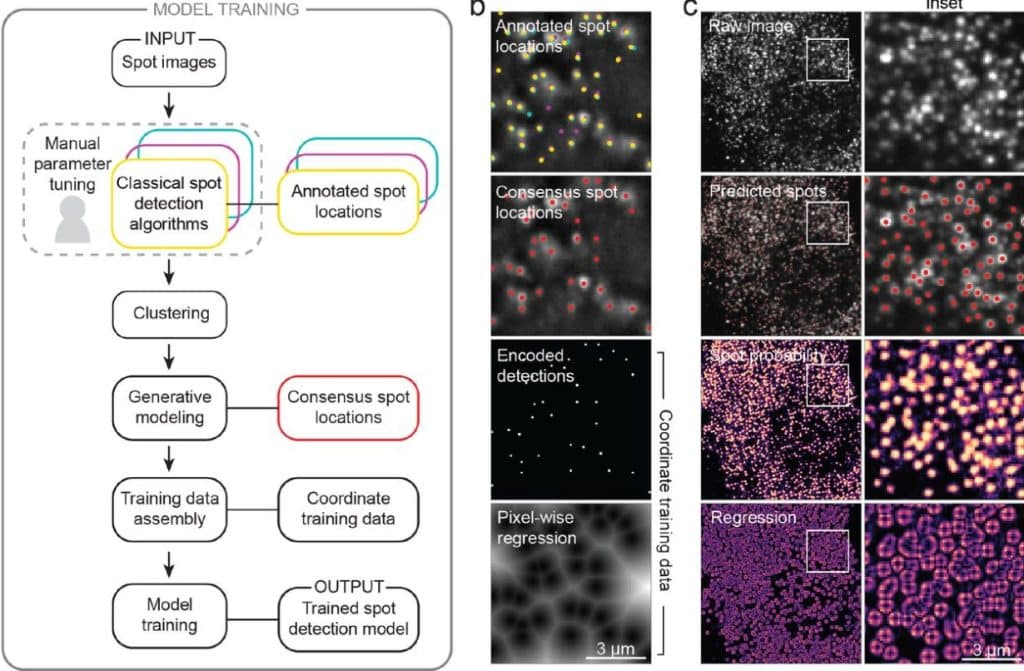
Accurate single-molecule spot detection for image-based spatial transcriptomics with weakly supervised deep learning
Emily Laubscher, Xuefei (Julie) Wang, Nitzan Razin, Tom Dougherty, Rosalind J. Xu, Lincoln Ombelets, Edward Pao, William Graf, Jeffrey R. Moffitt, Yisong Yue, David Van Valen
Generative adversarial networks in cell microscopy for image augmentation. A systematic review
Duway Nicolas Lesmes-Leon, Andreas Dengel, Sheraz Ahmed
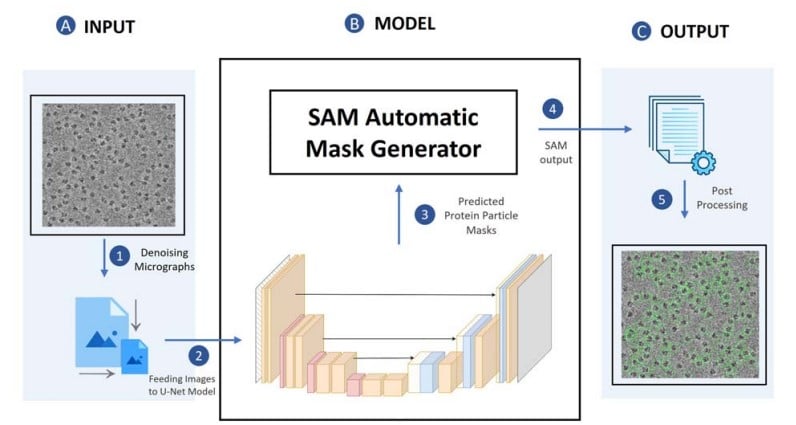
Accurate cryo-EM protein particle picking by integrating the foundational AI image segmentation model and specialized U-Net
Rajan Gyawali, Ashwin Dhakal, Liguo Wang, Jianlin Cheng
OneFlowTraX: A User-Friendly Software for Super-Resolution Analysis of Single-Molecule Dynamics and Nanoscale Organization
Leander Rohr, Alexandra Ehinger, Luiselotte Rausch, Nina Glöckner Burmeister, Alfred J. Meixner, Julien Gronnier, Klaus Harter, Birgit Kemmerling, Sven zur Oven-Krockhaus
SegmentAnything helps microscopy images based automatic and quantitative organoid detection and analysis
Xiaodan Xing, Chunling Tang, Yunzhe Guo, Nicholas Kurniawan, Guang Yang
Kartezio: Evolutionary Design of Explainable Pipelines for Biomedical Image Analysis
Kévin Cortacero, Brienne McKenzie, Sabina Müller, Roxana Khazen, Fanny Lafouresse, Gaëlle Corsaut, Nathalie Van Acker, François-Xavier Frenois, Laurence Lamant, Nicolas Meyer, Béatrice Vergier, Dennis G. Wilson, Hervé Luga, Oskar Staufer, Michael L. Dustin, Salvatore Valitutti, Sylvain Cussat-Blanc
Biomedical image analysis competitions: The state of current participation practice
Matthias Eisenmann, Annika Reinke, Vivienn Weru, Minu Dietlinde Tizabi, Fabian Isensee, Tim J. Adler, Patrick Godau, Veronika Cheplygina, Michal Kozubek, Sharib Ali, Anubha Gupta, Jan Kybic, Alison Noble, Carlos Ortiz de Solórzano, Samiksha Pachade, Caroline Petitjean, Daniel Sage, Donglai Wei, Elizabeth Wilden, Deepak Alapatt, Vincent Andrearczyk, Ujjwal Baid, Spyridon Bakas, Niranjan Balu, Sophia Bano , et al. (331 additional authors not shown)


 (No Ratings Yet)
(No Ratings Yet)