Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 9 February 2024
Here is a curated selection of preprints published recently. In this post, we focus specifically on new tools for bioimage analysis and data management.
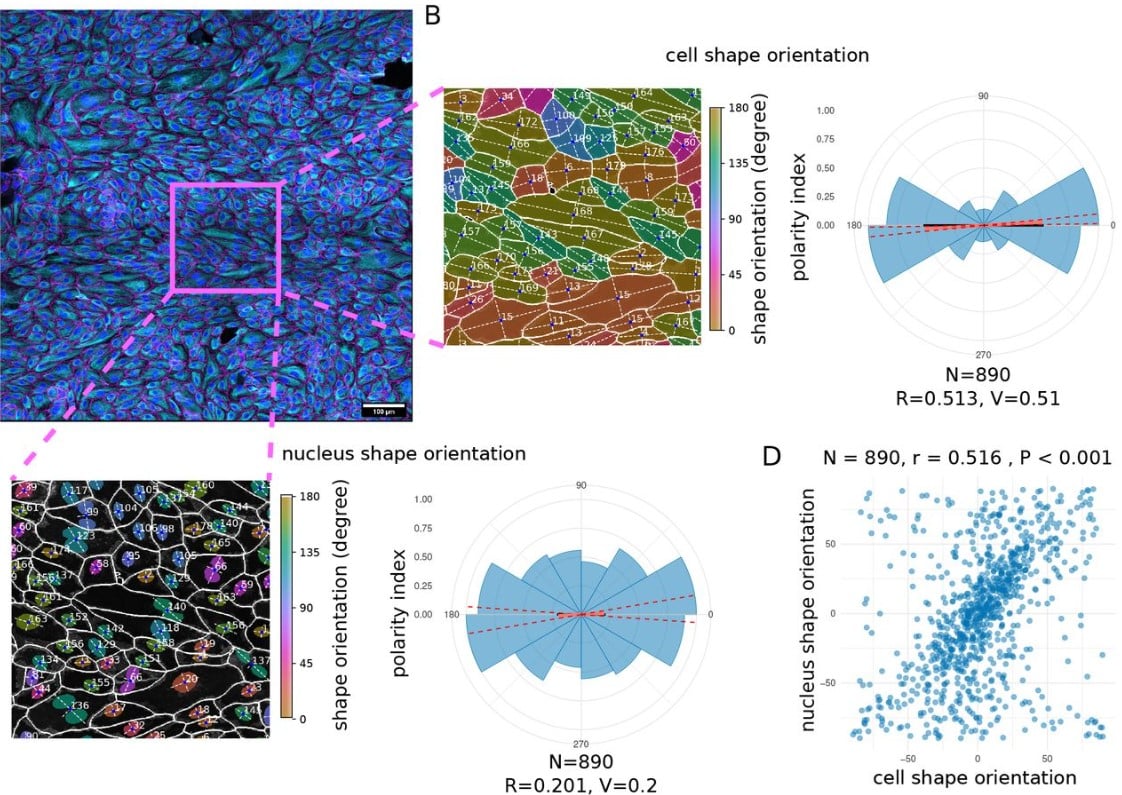
Polarity-JaM: An image analysis toolbox for cell polarity, junction and morphology quantification
Wolfgang Giese, Jan Philipp Albrecht, Olya Oppenheim, Emir Bora Akmeriç, Julia Kraxner, Deborah Schmidt, Kyle Harrington, Holger Gerhardt
A new protocol for multispecies bacterial infections in zebrafish and their monitoring through automated image analysis
Désirée A. Schmitz, Tobias Wechsler, Hongwei Bran Li, Bjoern Menze, Rolf Kümmerli
NIEND: Neuronal Image Enhancement through Noise Disentanglement
Zuo-Han Zhao, Yufeng Liu
Pseudo-spectral angle mapping for automated pixel-level analysis of highly multiplexed tissue image data
Madeleine S. Durkee, Junting Ai, Gabriel Casella, Thao Cao, Anthony Chang, Ariel Halper-Stromberg, Bana Jabri, Marcus R. Clark, Maryellen L. Giger
Self-supervision advances morphological profiling by unlocking powerful image representations
Vladislav Kim, Nikolaos Adaloglou, Marc Osterland, Flavio M. Morelli, Marah Halawa, Tim König, David Gnutt, Paula A. Marin Zapata
GPT-4V exhibits human-like performance in biomedical image classification
Wenpin Hou, Zhicheng Ji
Image processing tools for petabyte-scale light sheet microscopy data
Xiongtao Ruan, Matthew Mueller, Gaoxiang Liu, Frederik Görlitz, Tian-Ming Fu, Daniel E. Milkie, Joshua Lillvis, Alison Killilea, Eric Betzig, Srigokul Upadhyayula
An image segmentation pipeline optimized for human microglia uncovers sources of morphological diversity in Alzheimer’s disease
Robert M De Jager, Annie J Lee, Alina Sigalov, Mariko Taga
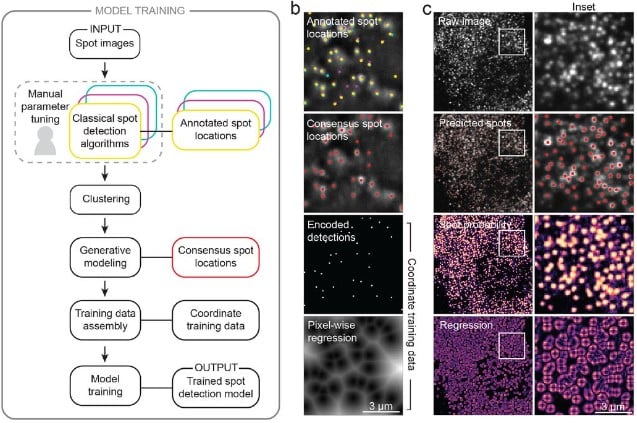
Accurate single-molecule spot detection for image-based spatial transcriptomics with weakly supervised deep learning
Emily Laubscher, Xuefei (Julie) Wang, Nitzan Razin, Tom Dougherty, Rosalind J. Xu, Lincoln Ombelets, Edward Pao, William Graf, Jeffrey R. Moffitt, Yisong Yue, David Van Valen
When the pen is mightier than the sword: semi-automatic 2 and 3D image labelling
Réka Hollandi, David Bauer, Akos Diosdi, Bálint Schrettner, Timea Toth, Dominik Hirling, Gábor Hollandi, Maria Harmati, József Molnár, Peter Horvath
The Brain Image Library: A Community-Contributed Microscopy Resource for Neuroscientists
Mariah Kenney, Iaroslavna Vasylieva, Greg Hood, Ivan Cao-Berg, Luke Tuite, Rozita Laghaei, Alan M. Watson, Alexander J. Ropelewski
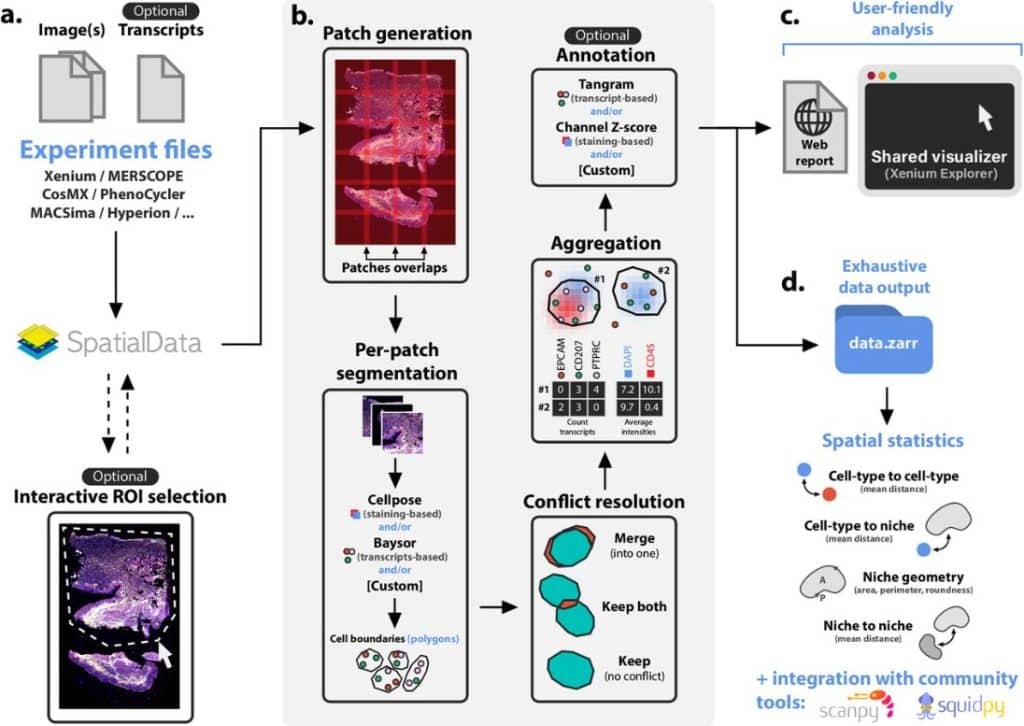
Sopa: a technology-invariant pipeline for analyses of image-based spatial-omics
Quentin Blampey, Kevin Mulder, Charles-Antoine Dutertre, Margaux Gardet, Fabrice André, Florent Ginhoux, Paul-Henry Cournède
CryoVirusDB: A Labeled Cryo-EM Image Dataset for AI-Driven Virus Particle Picking
Rajan Gyawali, Ashwin Dhakal, Liguo Wang, Jianlin Cheng
Self-inspired learning to denoise for live-cell super-resolution microscopy
Liying Qu, Shiqun Zhao, Yuanyuan Huang, Xianxin Ye, Kunhao Wang, Yuzhen Liu, Xianming Liu, Heng Mao, Guangwei Hu, Wei Chen, Changliang Guo, Jiaye He, Jiubin Tan, Haoyu Li, Liangyi Chen, Weisong Zhao
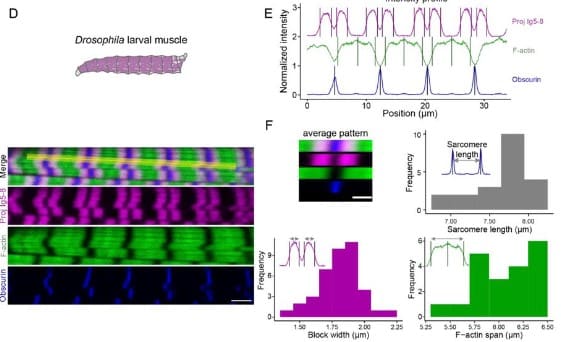
PatternJ: an ImageJ toolset for the automated and quantitative analysis of regular spatial patterns found in sarcomeres, axons, somites, and more
Mélina Baheux Blin, Vincent Loreau, Frank Schnorrer, Pierre Mangeol
Efficient stripe artefact removal by a variational method: application to light-sheet microscopy, FIB-SEM and remote sensing images
Niklas Rottmayer, Claudia Redenbach, Florian Fahrbach
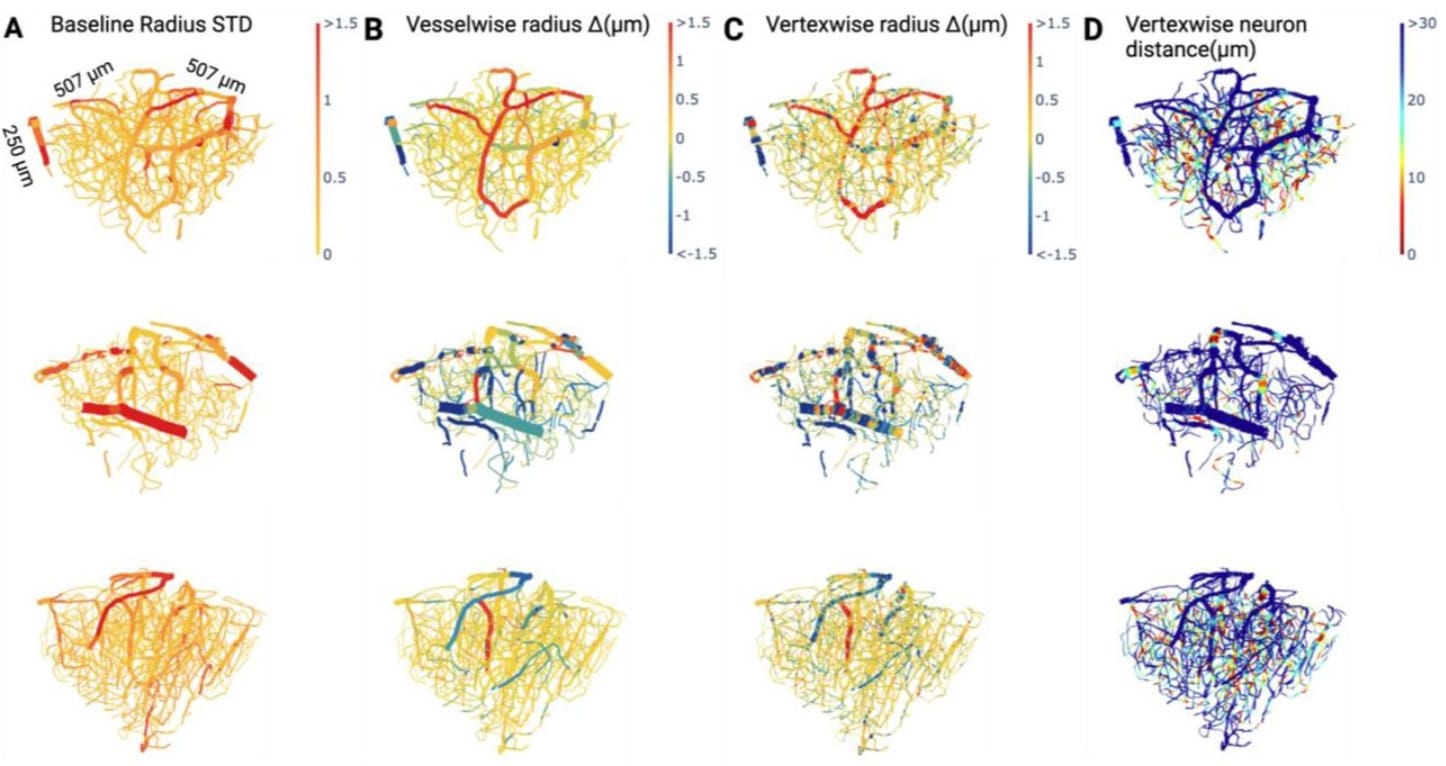
A Deep Learning Pipeline for Mapping in situ Network-level Neurovascular Coupling in Multi-photon Fluorescence Microscopy
Matthew Rozak, James Mester, Ahmadreza Attarpour, Adrienne Dorr, Maged Goubran, Bojana Stefanovic
Bridging the Gap: Integrating Cutting-edge Techniques into Biological Imaging with deepImageJ
Caterina Fuster-Barceló, Carlos García López de Haro, Estibaliz Gómez-de-Mariscal, Wei Ouyang, Jean-Christophe Olivo-Marin, Daniel Sage, Arrate Muñoz-Barrutia
CAMIL: Context-Aware Multiple Instance Learning for Cancer Detection and Subtyping in Whole Slide Images
Olga Fourkioti, Matt De Vries, Chris Bakal
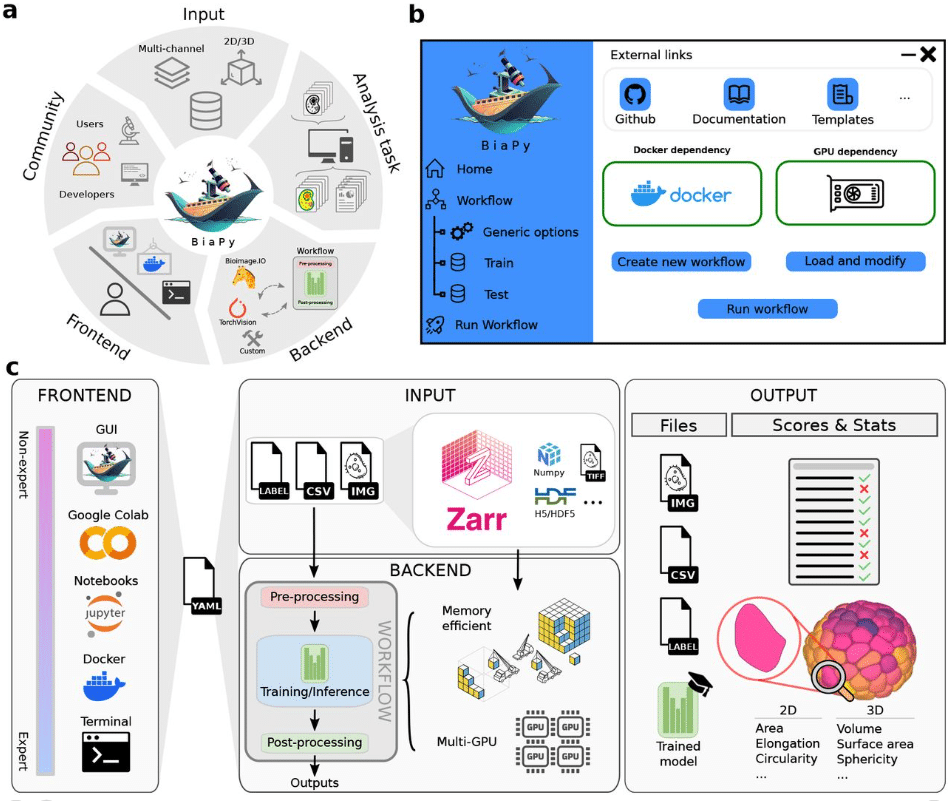
BiaPy: A unified framework for versatile bioimage analysis with deep learning
Daniel Franco-Barranco, Jesús A. Andrés-San Román, Ivan Hidalgo-Cenalmor, Lenka Backová, Aitor González-Marfil, Clément Caporal, Anatole Chessel, Pedro Gómez-Gálvez, Luis M. Escudero, Donglai Wei, Arrate Muñoz-Barrutia, Ignacio Arganda-Carreras
ExoJ: an ImageJ2/Fiji plugin for automated spatiotemporal detection and analysis of exocytosis
Junjun Liu, Frederik Johannes Verweij, Guillaume Van Niel, Thierry Galli, Lydia Danglot, Philippe Bun
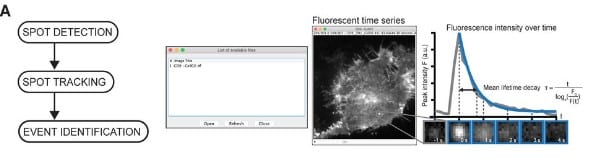
Figure extracted from Liu, et al. The image is made available under a CC-BY-NC 4.0 International license.
OneFlowTraX: A User-Friendly Software for Super-Resolution Analysis of Single-Molecule Dynamics and Nanoscale Organization
Leander Rohr, Alexandra Ehinger, Luiselotte Rausch, Nina Glöckner Burmeister, Alfred J. Meixner, Julien Gronnier, Klaus Harter, Birgit Kemmerling, Sven zur Oven-Krockhaus
MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography
Lorenz Lamm, Simon Zufferey, Ricardo D. Righetto, Wojciech Wietrzynski, Kevin A. Yamauchi, Alister Burt, Ye Liu, Hanyi Zhang, Antonio Martinez-Sanchez, Sebastian Ziegler, Fabian Isensee, Julia A. Schnabel, Benjamin D. Engel, Tingying Peng
A Novel, Open-Source Virtual Reality Platform for Dendritic Spine Analysis
Marike L Reimer, Sierra D Kauer, Curtis A Benson, Jared F King, Siraj Patwa, Sarah Feng, Maile A Estacion, Lakshmi Bangalore, Stephen Waxman, Andrew M Tan
Piscis: a novel loss estimator of the F1 score enables accurate spot detection in fluorescence microscopy images via deep learning
Zijian Niu, Aoife O’Farrell, Jingxin Li, Sam Reffsin, Naveen Jain, Ian Dardani, Yogesh Goyal, Arjun Raj
ThirdPeak: A flexible tool designed for the robust analysis of two- and three-dimensional (single-molecule) tracking data
Thomas Müller, Elisabeth Meiser, Markus Engstler
Enabling Global Image Data Sharing in the Life Sciences
Peter Bajcsy, Sreenivas Bhattiprolu, Katy Boerner, Beth A Cimini, Lucy Collinson, Jan Ellenberg, Reto Fiolka, Maryellen Giger, Wojtek Goscinski, Matthew Hartley, Nathan Hotaling, Rick Horwitz, Florian Jug, Anna Kreshuk, Emma Lundberg, Aastha Mathur, Kedar Narayan, Shuichi Onami, Anne L. Plant, Fred Prior, Jason Swedlow, Adam Taylor, Antje Keppler
AnNoBrainer, an Automated Annotation of Mouse Brain Images using Deep Learning
Roman Peter, Petr Hrobar, Josef Navratil, Martin Vagenknecht, Jindrich Soukup, Keiko Tsuji, Nestor X. Barrezueta, Anna C. Stoll, Renee C. Gentzel, Jonathan A. Sugam, Jacob Marcus, Danny A. Bitton
A systematic evaluation of computation methods for cell segmentation
Yuxing Wang, Junhan Zhao, Hongye Xu, Cheng Han, Zhiqiang Tao, Dongfang Zhao, Dawei Zhou, Gang Tong, Dongfang Liu, Zhicheng Ji
PCP Auto Count: A Novel Fiji/ImageJ plug-in for automated quantification of planar cell polarity and cell counting
Kendra L. Stansak, Luke D. Baum, Sumana Ghosh, Punam Thapa, Vineel Vanga, Bradley J. Walters
Spotiflow: accurate and efficient spot detection for imaging-based spatial transcriptomics with stereographic flow regression
Albert Dominguez Mantes, Antonio Herrera, Irina Khven, Anjalie Schlaeppi, Eftychia Kyriacou, Georgios Tsissios, Can Aztekin, Joachim Lingner, Gioele La Manno, Martin Weigert
SC-Track: a robust cell tracking algorithm for generating accurate single-cell lineages from diverse cell segmentations
Chengxin Li, Shuang Shuang Xie, Jiaqi Wang, Septavera Sharvia, Kuan Yoow Chan
CellTracksColab—A platform for compiling, analyzing, and exploring tracking data
Guillaume Jacquemet
Automated neuropil segmentation of fluorescent images for Drosophila brains
Kai-Yi Hsu, Chi-Tin Shih, Nan-Yow Chen, Chung-Chuan Lo
Unleashing the Infinity Power of Geometry: A Novel Geometry-Aware Transformer (GOAT) for Whole Slide Histopathology Image Analysis
Mingxin Liu, Yunzan Liu, Pengbo Xu, Jiquan Ma


 (No Ratings Yet)
(No Ratings Yet)