Microscopy preprints – bioimage analysis tools
Posted by FocalPlane, on 17 November 2023
Here is a curated selection of preprints published recently. In this post, we focus specifically on new tools for bioimage analysis and data management.
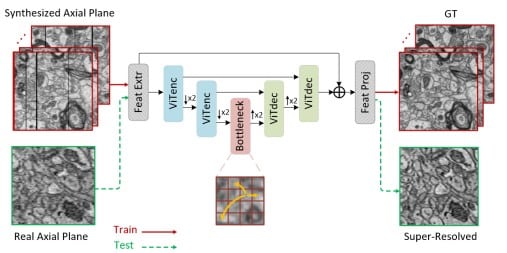
Self-Supervised Super-Resolution Approach for Isotropic Reconstruction of 3D Electron Microscopy Images from Anisotropic Acquisition
Mohammad Khateri, Morteza Ghahremani, Alejandra Sierra, Jussi Tohka
DynaMight: estimating molecular motions with improved reconstruction from cryo-EM images
Johannes Schwab, Dari Kimanius, Alister Burt, Tom Dendooven, Sjors H.W. Scheres
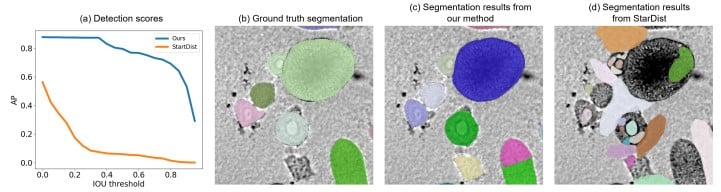
Unsupervised Learning of Object-Centric Embeddings for Cell Instance Segmentation in Microscopy Images
Steffen Wolf, Manan Lalit, Henry Westmacott, Katie McDole, Jan Funke

Towards quantitative super-resolution microscopy: Molecular maps with statistical guarantees
Katharina Proksch, Frank Werner, Jan Keller-Findeisen, Haisen Ta, Axel Munk
SegmentAnything helps microscopy images based automatic and quantitative organoid detection and analysis
Xiaodan Xing, Chunling Tang, Yunzhe Guo, Nicholas Kurniawan, Guang Yang
PC-bzip2: a phase-space continuity enhanced lossless compression algorithm for light field microscopy data
Changqing Su, Zihan Lin, You Zhou, Shuai Wang, Yuhan Gao, Chenggang Yan, Bo Xiong
Deep learning-based aberration compensation improves contrast and resolution in fluorescence microscopy
Min Guo, Yicong Wu, Yijun Su, Shuhao Qian, Eric Krueger, Ryan Christensen, Grant Kroeschell, Johnny Bui, Matthew Chaw, Lixia Zhang, Jiamin Liu, Xuekai Hou, Xiaofei Han, Xuefei Ma, Alexander Zhovmer, Christian Combs, Mark Moyle, Eviatar Yemini, Huafeng Liu, Zhiyi Liu, Patrick La Riviere, Daniel Colón-Ramos, Hari Shroff

Fine-tuning TrailMap: The utility of transfer learning to improve the performance of deep learning in axon segmentation of light-sheet microscopy images
Marjolein Oostrom, Michael A. Muniak, Rogene M. Eichler West, Sarah Akers, Paritosh Pande, Moses Obiri, Wei Wang, Kasey Bowyer, Zhuhao Wu, Lisa M. Bramer, Tianyi Mao, Bobbie Jo Webb-Robertson
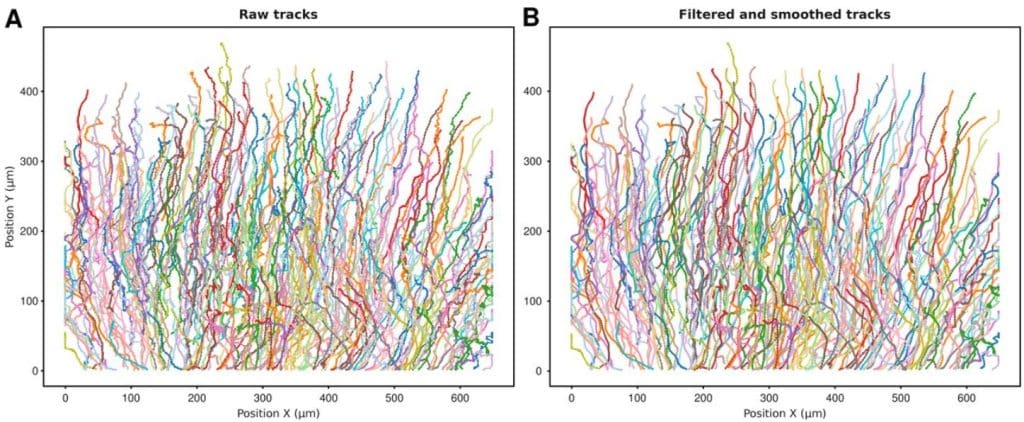
CellTracksColab—A platform for compiling, analyzing, and exploring tracking data
Guillaume Jacquemet
Digitalized organoids: integrated pipeline for 3D high-speed analysis of organoid structures using multilevel segmentation and cellular topology
Hui Ting Ong, Esra Karatas, Gianluca Grenci, Florian Dilasser, Saburnisha Binte Mohamad Raffi, Damien Blanc, Titouan Poquillon, Elise Drimaracci, Dimitri Mikec, Cora Thiel, Oliver Ullrich, Victor Racine, Anne Beghin
Fast and universal single molecule localization using multi-dimensional point spread functions
Mengfan Li, Wei Shi, Sheng Liu, Shuang Fu, Yue Fei, Lulu Zhou, Yiming Li
Fokker-Planck analysis of superresolution microscopy images
Mario Annunziato, Alfio Borzì
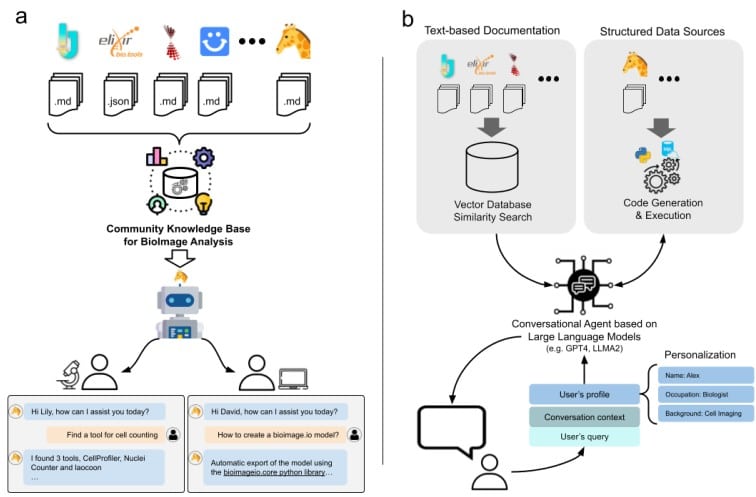
BioImage.IO Chatbot: A Personalized Assistant for BioImage Analysis Augmented by Community Knowledge Base
Wanlu Lei, Caterina Fuster-Barceló, Arrate Muñoz-Barrutia, Wei Ouyang
I3D:bio’s OMERO training material: Re-usable, adjustable, multi-purpose slides for local user training
Schmidt, Christian, Bortolomeazzi, Michele, Boissonnet, Tom, Fortmann-Grote, Carsten, Dohle, Julia, Zentis, Peter, Kandpal, Niraj,Kunis, Susanne, Zobel, Thomas, Weidtkamp-Peters, Stefanie, Ferrando-May, Elisa
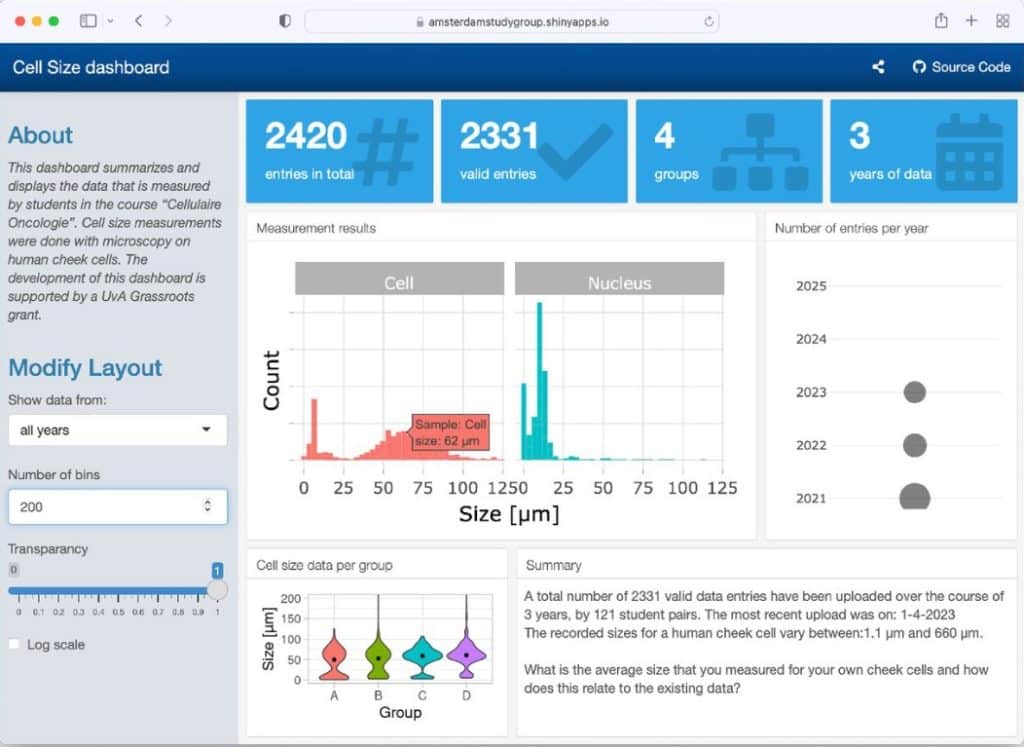
Studentsourcing – aggregating and re-using data from a practical cell biology course
Joachim Goedhart
Computational Tools for the Analysis of Meiotic Prophase I Images
James Hugh Crichton, Ian R Adams

ConfluentFUCCI for fully-automated analysis of cell-cycle progression in a highly dense collective of migrating cells
Leo Goldstien, Yael Lavi, Lior Atia
HiTIPS: High-Throughput Image Processing Software for the Study of Nuclear Architecture and Gene Expression
Adib Keikhosravi, Faisal Almansour, Christopher H. Bohrer, Nadezda A. Fursova, Krishnendu Guin, Varun Sood, Tom Misteli, Daniel R. Larson, Gianluca Pegoraro
EMDB – the Electron Microscopy Data Bank
Jack Turner, The wwPDB Consortium
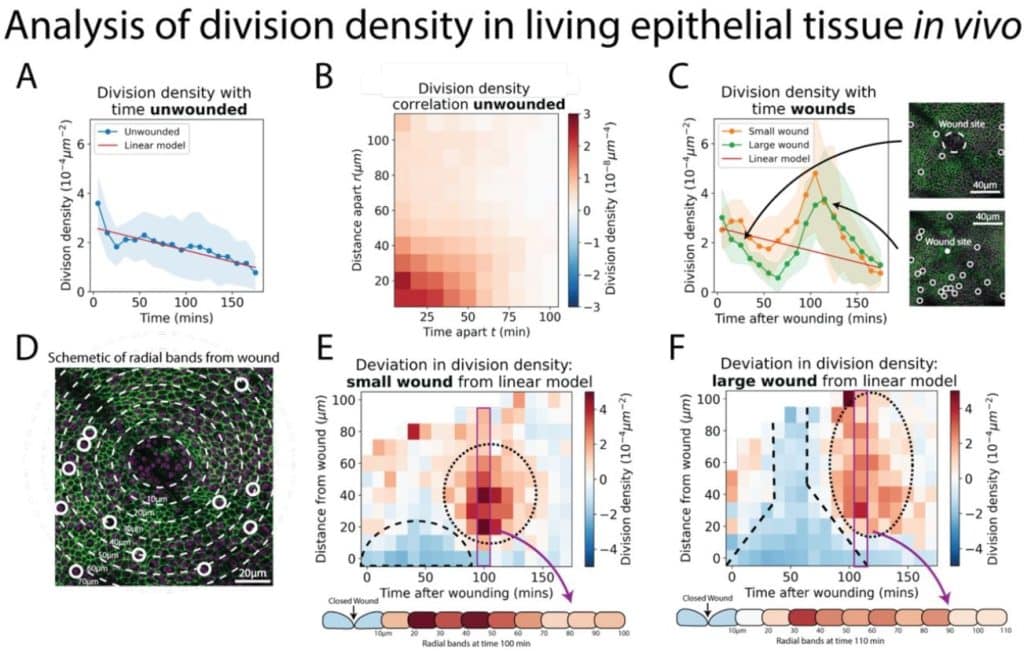
Deep learning for rapid analysis of cell divisions in vivo during epithelial morphogenesis and repair
Jake Turley, Isaac V. Chenchiah, Paul Martin, Tanniemola B. Liverpool, Helen Weavers
Virtual tissue microstructure reconstruction across species using generative deep learning
Nicolás Bettancourt, Cristian Pérez, Valeria Candia, Pamela Guevara, Yannis Kalaidzidis, Marino Zerial, Fabián Segovia-Miranda, Hernán Morales-Navarrete
NIEND: Neuronal Image Enhancement through Noise Disentanglement
Zuo-Han Zhao, Yufeng Liu
FICTURE: Scalable segmentation-free analysis of submicron resolution spatial transcriptomics
Yichen Si, ChangHee Lee, Yongha Hwang, Jeong H. Yun, Weiqiu Cheng, Chun-Seok Cho, Miguel Quiros, Asma Nusrat, Weizhou Zhang, Goo Jun, Sebastian Zöllner, Jun Hee Lee, Hyun Min Kang
A Bayesian Framework for Cryo-EM Heterogeneity Analysis using Regularized Covariance Estimation
Marc Aurèle Gilles, Amit Singer


 (No Ratings Yet)
(No Ratings Yet)