A general strategy to develop fluorogenic polymethine dyes for bioimaging
Posted by Annabell Martin, on 6 December 2023
Annabell Martin1,2 and Pablo Rivera-Fuentes1*
1) University of Zurich; Department of Chemistry, CH-8057 Zurich, Switzerland.
2) École Polytechnique Fédérale de Lausanne (EPFL), Institute of Chemical Sciences and Engineering, CH-1015 Lausanne, Switzerland.
* Corresponding author
https://www.nature.com/articles/s41557-023-01367-y
Rivera-Fuentes Lab: https://www.locbp.com @RiveraChemLab
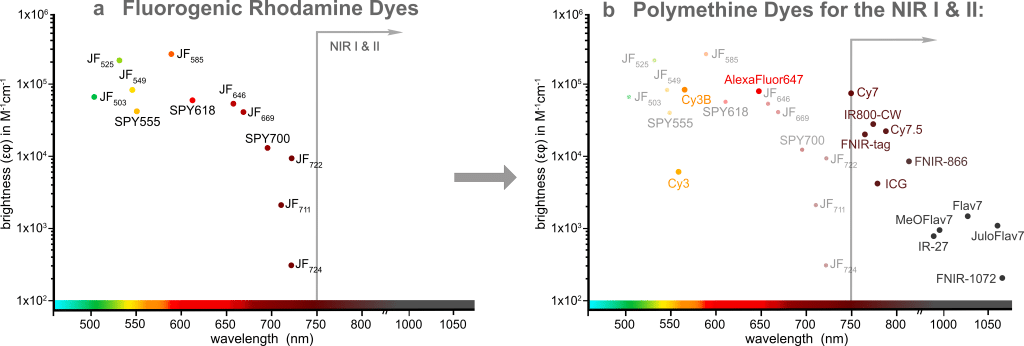
Fluorescence imaging is an invaluable tool to study biological processes and further progress depends on the development of novel advanced fluorescent probes. Traditional fluorophores often suffer from low cell permeability and high background fluorescence that requires extensive washing steps and therefore limits their use in microscopy experiments. Over the last decade, several fluorogenic rhodamine dyes have been developed: The JaneliaFluor® dyes1,2 and the SPYTM dyes3, among others. These dyes are non-fluorescent in solution but become highly fluorescent when bound to their specific target. They have been utilized extensively in no-wash, multi-color live cell imaging experiments, but despite the tremendous success of these dyes in the visible spectral window, this rhodamine-based fluorogenicity strategy cannot be applied to the NIR I and II (Figure 1a).
We decided to tackle this problem by harnessing another dye scaffold, which is called polymethine dyes. These dyes are among the most-used fluorophores for cell, tissue, and whole-organism imaging due to their simple and highly modular synthesis, high extinction coefficients, biocompatibility, and most importantly tunable emission wavelength spanning from the green to the shortwave infrared wavelengths, thus reaching the NIR I and II imaging windows. If you’ve ever performed super-resolution microscopy (in particular STORM) experiments, you may have heard of or already used AlexaFluor® 6474; or if you’re working in the in vivo imaging field, you are familiar with the clinically approved Indocyanine Green (ICG)5 as well. These are polymethine dyes (Figure 1b).
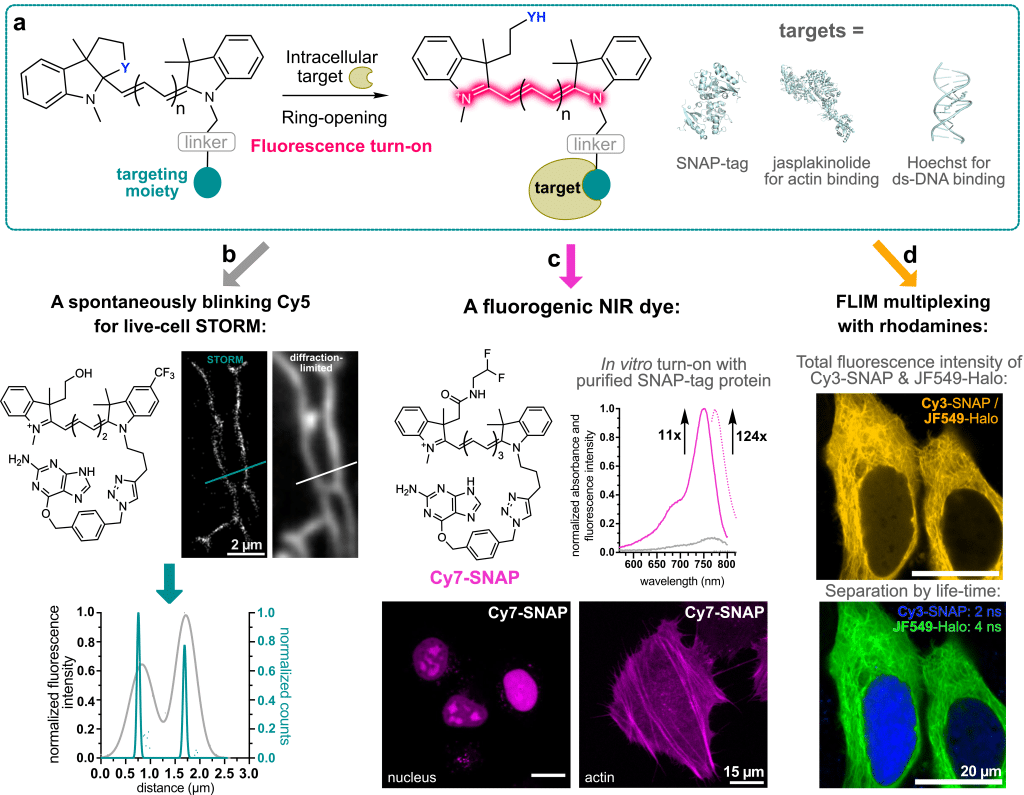
In contrast to rhodamine dyes, this polymethine dye class does not have a built-in cyclization equilibrium that can be leveraged to make the dyes fluorogenic. In our laboratory, we recently developed a strategy to overcome this challenge: We used chemistry heuristics to incorporate a ring-closing moiety into the classical dye scaffold that disrupts the conjugated system and makes the fluorophore non-fluorescent in solution6. Only upon binding to its specific intracellular target, the ring opens and renders the molecule fluorescent. This binding-induced fluorescence turn-on can be achieved with self-labeling protein tags, such as SNAP-tag, as well as with small-molecule targeting ligands (Figure 2a).
Most excitingly, using polymethines as a scaffold for fluorogenic dyes opens up a lot of new avenues for bioimaging. Despite this being only the beginning, we were already able to demonstrate that:
- With a hydroxy ethyl ring-closing nucleophile – very similar to HMSiR – a spontaneously blinking Cy5 dye can be created. This dye can be used in live-cell STORM experiments in standard growth medium without the need for blinking or anti-fading agents (Figure 2b).
- The spectral properties of polymethine dyes can be easily extended into the NIR by increasing the number of polymethine units. Therefore, by simply varying commercially available linker, we were able to generate a fluorogenic Cy7-SNAP dye (Figure 2c). Furthermore, binding to the protein surface enhances the quantum yield of the Cy7 significantly to 33% and thus provides access to very bright and photostable polymethine dyes.
- These dyes can be used in multiplexing experiments together with rhodamine dyes. Due to their vastly different dye scaffolds, rhodamines and polymethine dyes have very different fluorescence lifetimes. This can be leveraged to use dyes with the same spectral properties, e.g., a Cy3 and a JF549, and still separate the fluorescence signal by lifetime (Figure 2d).
Our fluorogenicity approach can be leveraged to create many bright and photostable near-infrared and shortwave-infrared probes and sensors. The next step is to test these dyes in vivo to evaluate their ability to label self-labeling proteins in tissue and to compare these probes with genetically encoded NIR-fluorescent proteins. We envision that our simple, yet general, method to develop fluorogenic polymethine probes will facilitate the creation of NIR probes with improved photophysical and physicochemical properties for the next generation of chemigenetic imaging probes.
Citations:
1. Grimm, J. B. et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 14, 987–994 (2017).
2. Grimm, J. B. et al. A general method to optimize and functionalize red-shifted rhodamine dyes. Nat. Methods 17, 815–821 (2020).
3. Wang, L. et al. A general strategy to develop cell permeable and fluorogenic probes for multicolour nanoscopy. Nat. Chem. 12, 165–172 (2020).
4. Lelek, M. et al. Single-molecule localization microscopy. Nat. Rev. Methods Primer 1, 39 (2021).
5. Alander, J. T. et al. A review of Indocyanine Green fluorescent imaging in surgery. Int. J. Biomed. Imag. 2012, 1–26 (2012).
6. Martin, A. & Rivera-Fuentes, P. A general strategy to develop fluorogenic polymethine dyes for bioimaging. Nat. Chem. in press, https://doi.org/10.1038/s41557-023-01367-y.


 (No Ratings Yet)
(No Ratings Yet)