Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 22 March 2024
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis.
A Call for FAIR and Open-Access Training Materials to advance Bioimage Analysis
Robert Haase, Christian Tischer, Peter Bankhead, Kota Miura, and Beth Cimini
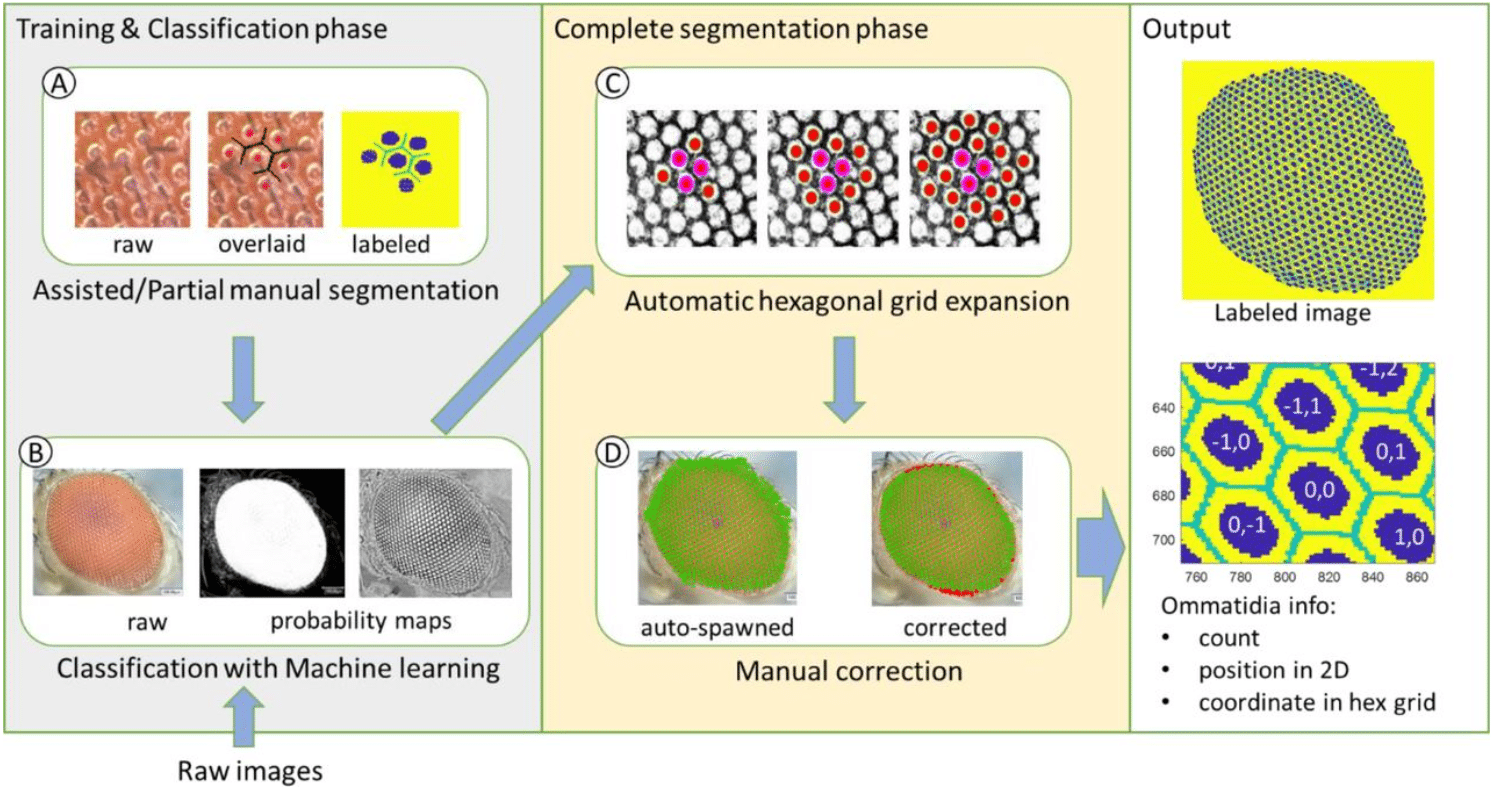
EyeHex toolbox for complete segmentation of ommatidia in fruit fly eyes
Huy Tran, Nathalie Dostatni, Ariane Ramaekers
Miffi: Improving the accuracy of CNN-based cryo-EM micrograph filtering with fine-tuning and Fourier space information
Da Xu, Nozomi Ando
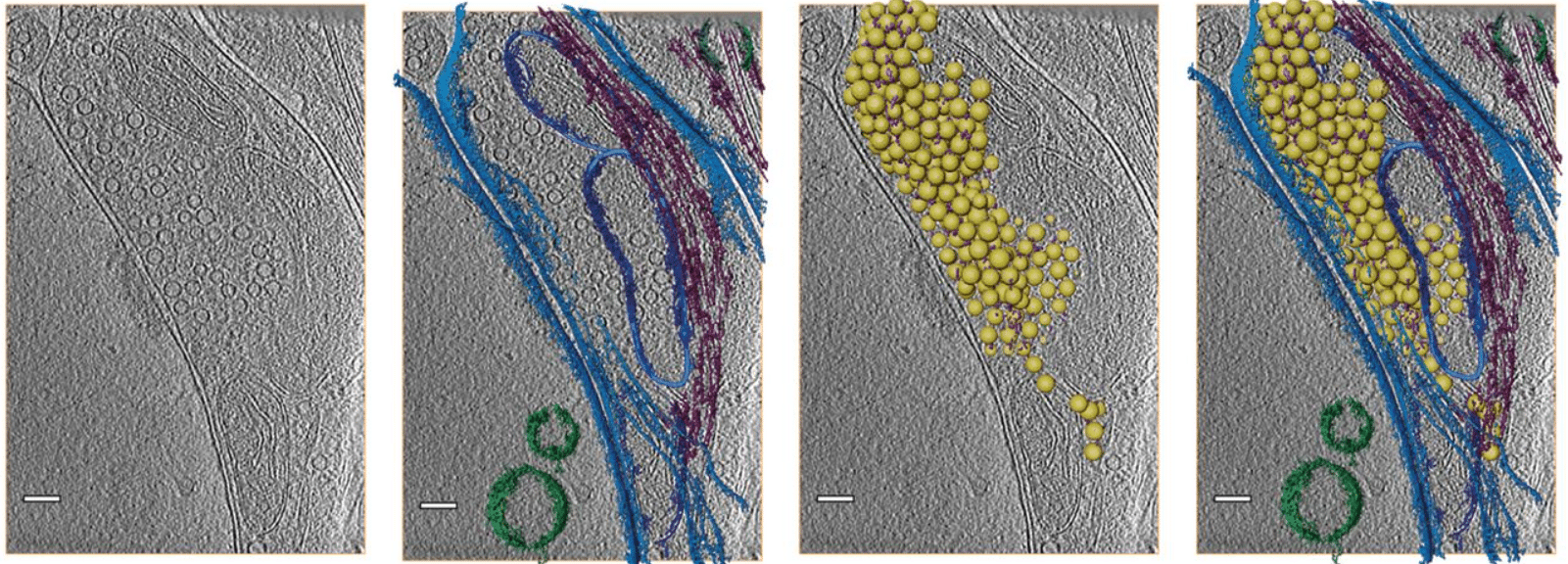
CryoVesNet: A Dedicated Framework for Synaptic Vesicle Segmentation in Cryo Electron Tomograms
Amin Khosrozadeh, Raphaela Seeger, Guillaume Witz, Julika Radecke, Jakob B. Sørensen, Benoît Zuber
QUAL-IF-AI: Quality Control of Immunofluorescence Images using Artificial Intelligence
Madhavi Dipak Andhari, Giulia Rinaldi, Pouya Nazari, Gautam Shankar, Nikolina Dubroja, Johanna Vets, Tessa Ostyn, Maxime Vanmechelen, Brecht Decraene, Alexandre Arnould, Willem Mestdagh, Bart De Moor, Frederik De Smet, Francesca Bosisio, Asier Antoranz
The Berkeley Single Cell Computational Microscopy (BSCCM) Dataset
Henry Pinkard, Cherry Liu, Fanice Nyatigo, Daniel A. Fletcher, Laura Waller
No-Clean-Reference Image Super-Resolution: Application to Electron Microscopy
Mohammad Khateri, Morteza Ghahremani, Alejandra Sierra, Jussi Tohka
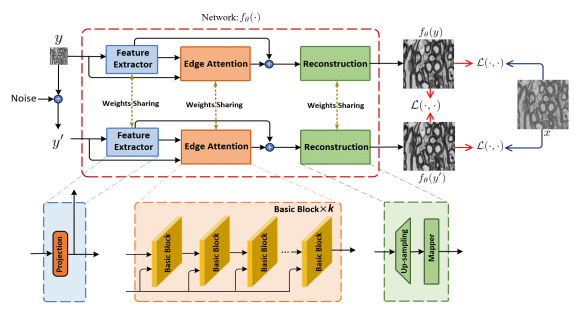
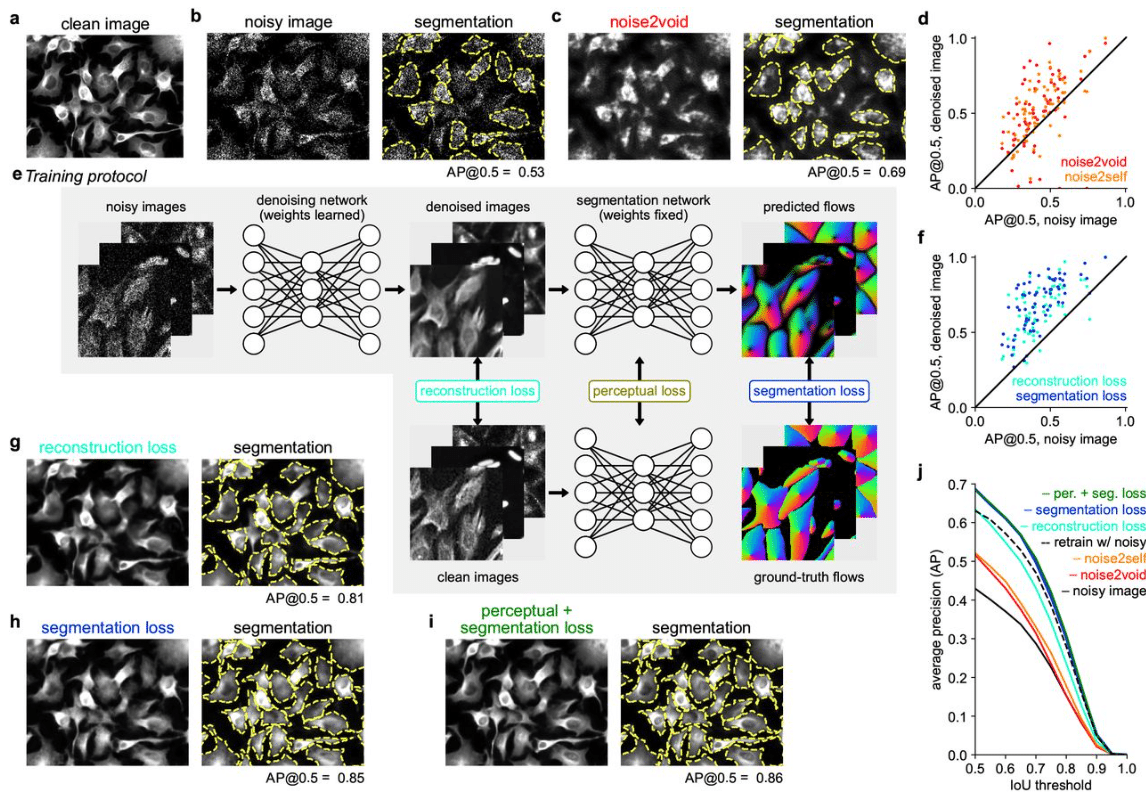
Cellpose3: one-click image restoration for improved cellular segmentation
Carsen Stringer, Marius Pachitariu
A deep learning-based toolkit for 3D nuclei segmentation and quantitative analysis in cellular and tissue context
Athul Vijayan, Tejasvinee Atul Mody, Qin Yu, Adrian Wolny, Lorenzo Cerrone, Soeren Strauss, Miltos Tsiantis, Richard S. Smith, Fred A. Hamprecht, Anna Kreshuk, Kay Schneitz
TomoNet: A streamlined cryoET software pipeline with automatic particle picking on flexible lattices
Hui Wang, Shiqing Liao, Xinye Yu, Jiayan Zhang, Z. Hong Zhou
Generative interpolation and restoration of images using deep learning for improved 3D tissue mapping
Saurabh Joshi, André Forjaz, Kyu Sang Han, Yu Shen, Daniel Xenes, Jordan Matelsky, Brock Wester, Arrate Munoz Barrutia, Ashley L. Kiemen, Pei-Hsun Wu, Denis Wirtz
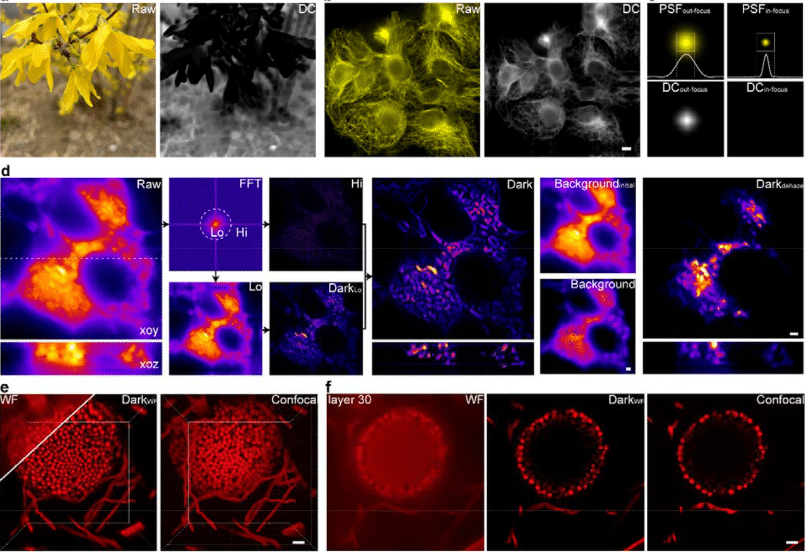
Dark-based Optical Sectioning assists Background Removal in Fluorescence Microscopy
Ruijie Cao, Yaning Li, Wenyi Wang, Guoxun Zhang, Gang Wang, Yu Sun, Wei Ren, Jing Sun, Yiwei Hou, Xinzhu Xu, Jiakui Hu, Yanye Lu, Changhui Li, Jiamin Wu, Meiqi Li, Junle Qu, Peng Xi
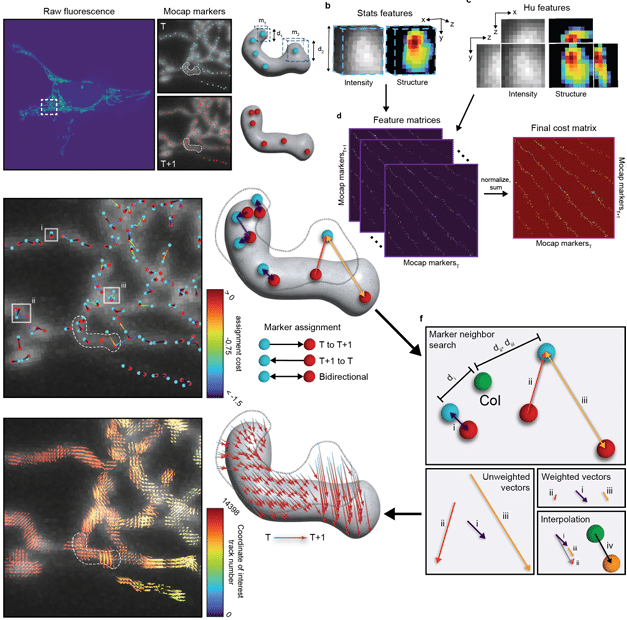
Nellie: Automated organelle segmentation, tracking, and hierarchical feature extraction in 2D/3D live-cell microscopy
Austin E. Y. T. Lefebvre, Gabriel Sturm, Ting-Yu Lin, Emily Stoops, Magdalena Preciado Lopez, Benjamin Kaufmann-Malaga, Kayley Hake
Celldetective: an AI-enhanced image analysis tool for unraveling dynamic cell interactions
Rémy Torro, Beatriz Dìaz-Bello, Dalia El Arawi, Lorna Ammer, Patrick Chames, Kheya Sengupta, Laurent Limozin
Empowering high-throughput high-content analysis of 3D tumor models: Open-source software for automated non-confocal image analysis
Noah Wiggin, Carson Cook, Mitchell Black, Ines Cadena, Salam Rahal-Arabi, Kaitlin Fogg
Insights into Cellular Evolution: Temporal Deep Learning Models and Analysis for Cell Image Classification
Xinran Zhao, Alexander Ruys de Perez, Elena S. Dimitrova, Melissa Kemp, Paul E. Anderson
Evaluating batch correction methods for image-based cell profiling
John Arevalo, Ellen Su, Robert van Dijk, Anne E. Carpenter, Shantanu Singh
Image processing tools for petabyte-scale light sheet microscopy data
Xiongtao Ruan, Matthew Mueller, Gaoxiang Liu, Frederik Görlitz, Tian-Ming Fu, Daniel E. Milkie, Joshua L. Lillvis, Alexander Kuhn, Chu Yi Aaron Herr, Wilmene Hercule, Marc Nienhaus, Alison N. Killilea, Eric Betzig, Srigokul Upadhyayula
An unsupervised deep learning framework encodes super-resolved image features to decode bacterial cell cycle
Juliette Griffié, Chen Zhang, Julien Denereaz, Thanh-An Pham, Gauthier Weissbart, Christian Sieben, Ambroise Lambert, Jan-Willem Veening, Suliana Manley
DeepSLICEM: Clustering CryoEM particles using deep image and similarity graph representations
Meghana V. Palukuri, Edward M. Marcotte
Accurate cryo-EM protein particle picking by integrating the foundational AI image segmentation model and specialized U-Net
Rajan Gyawali, Ashwin Dhakal, Liguo Wang, Jianlin Cheng
nERdy: network analysis of endoplasmic reticulum dynamics
Ashwin Samudre, Guang Gao, Ben Cardoen, Ivan Robert Nabi, Ghassan Hamarneh
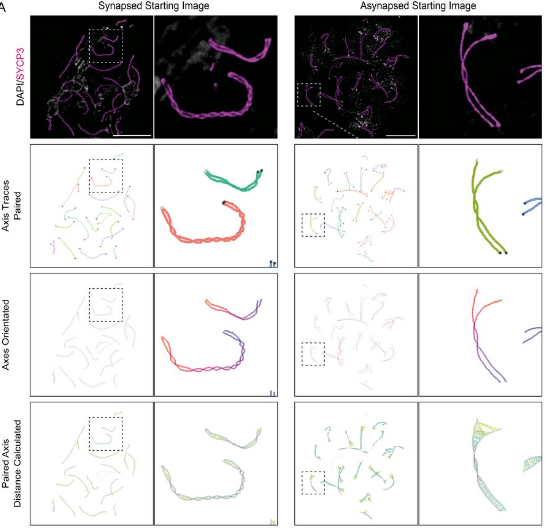
Computational Tools for the Analysis of Meiotic Prophase I Images
James H Crichton, Ian R Adams
MultiMatch: Geometry-Informed Colocalization in Multi-Color Super-Resolution Microscopy
Julia Naas, Giacomo Nies, Housen Li, Stefan Stoldt, Bernhard Schmitzer, Stefan Jakobs, Axel Munk


 (1 votes, average: 1.00 out of 1)
(1 votes, average: 1.00 out of 1)