Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 3 May 2024
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis.
FInCH: FIJI plugin for automated and scalable whole-image analysis of protein expression and cell morphology
Cody A. Lee, Carmen Sánchez Moreno, Alexander V. Badyaev
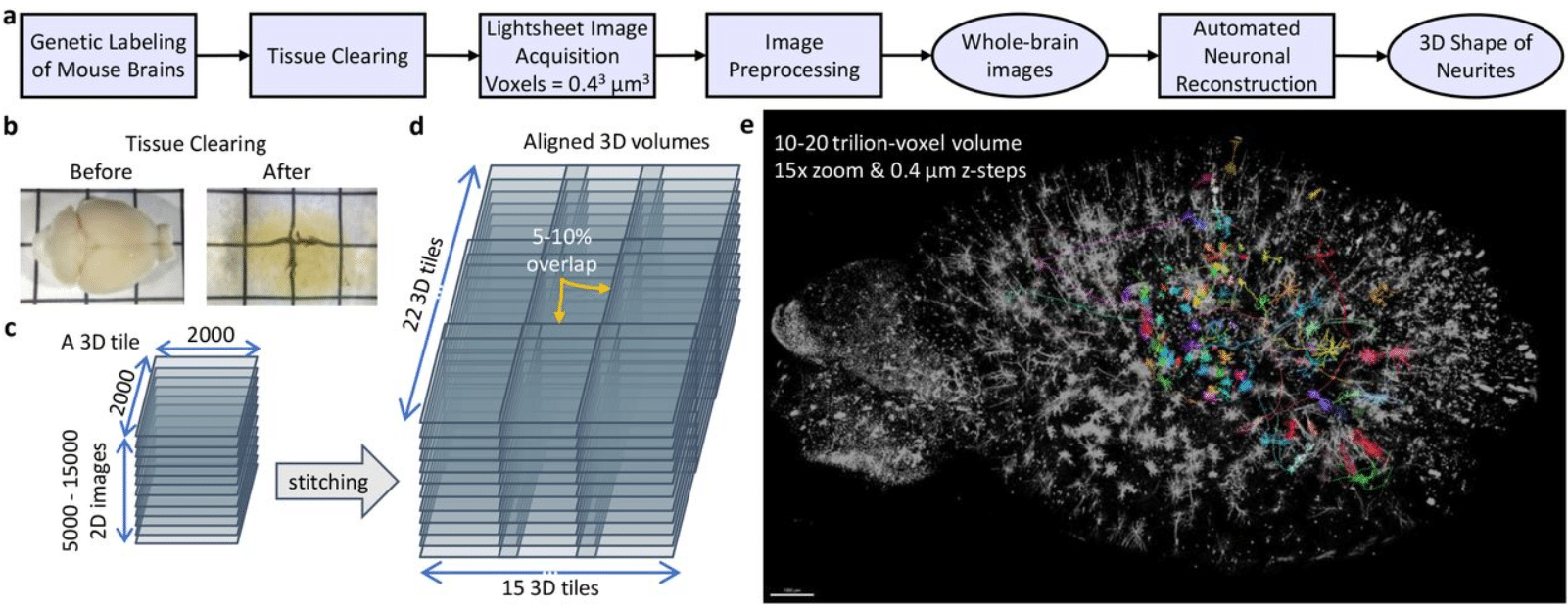
Gossamer: Scaling Image Processing and Reconstruction to Whole Brains
Karl Marrett, Keivan Moradi, Chris Sin Park, Ming Yan, Chris Choi, Muye Zhu, Masood Akram, Sumit Nanda, Qing Xue, Hyun-Seung Mun, Adriana Gutierez, Mitchell Rudd, Brian Zingg, Gabrielle Magat, Kathleen Wijaya, Hongwei Dong, X. William Yang, Jason Cong
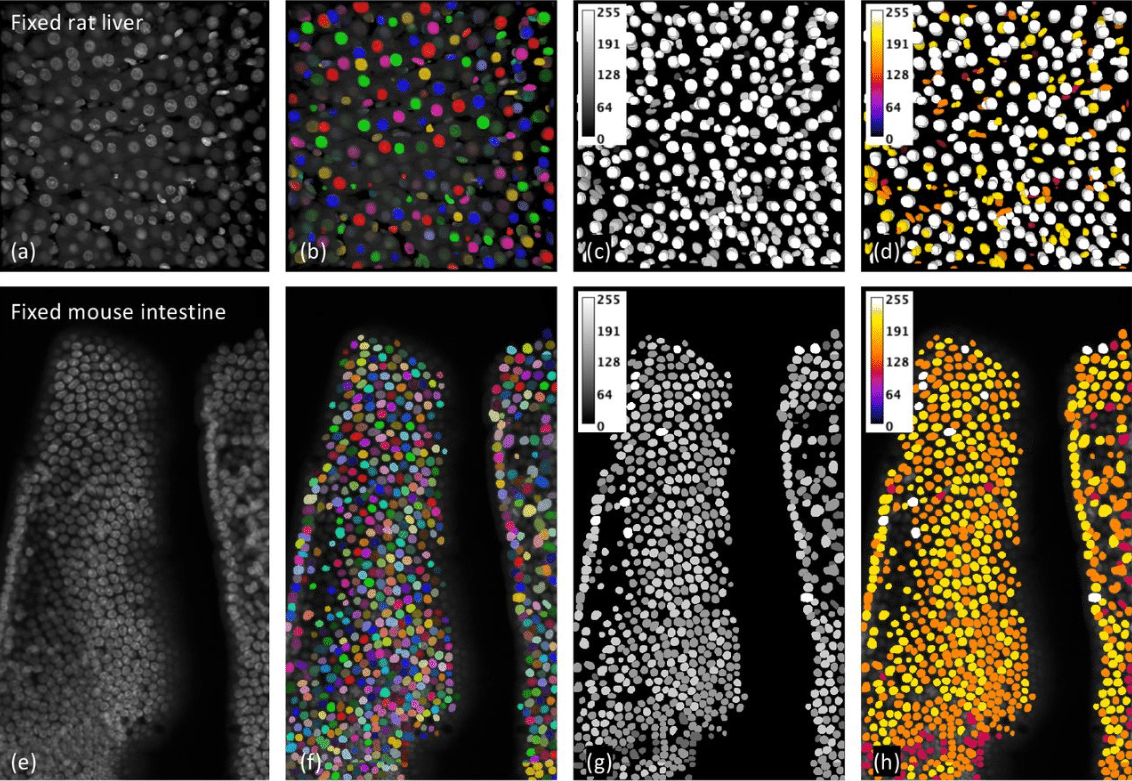
A Quantitative Metric of Confidence For Segmentation of Nuclei in Large Spatially Variable Image Volumes
Liming Wu, Alain Chen, Paul Salama, Kenneth W. Dunn, Seth Winfree, Edward J. Delp
StomaVision: stomatal trait analysis through deep learning
Ting-Li Wu, Po-Yu Chen, Xiaofei Du, Heiru Wu, Jheng-Yang Ou, Po-Xing Zheng, Yu-Lin Wu, Ruei-Shiuan Wang, Te-Chang Hsu, Chen-Yu Lin, Wei-Yang Lin, Ping-Lin Chang, Chin-Min Kimmy Ho, Yao-Cheng Lin
SPACe (Swift Phenotypic Analysis of Cells): an open-source, single cell analysis of Cell Painting data
Fabio Stossi, Pankaj K. Singh, Michela Marini, Kazem Safari, Adam T. Szafran, Alejandra Rivera Tostado, Christopher D. Candler, Maureen G. Mancini, Elina A. Mosa, Michael J. Bolt, Demetrio Labate, Michael A. Mancini
Analysis Pipeline to Quantify Uterine Gland Structural Variations
Sameed Khan, Adam Alessio, Ripla Arora
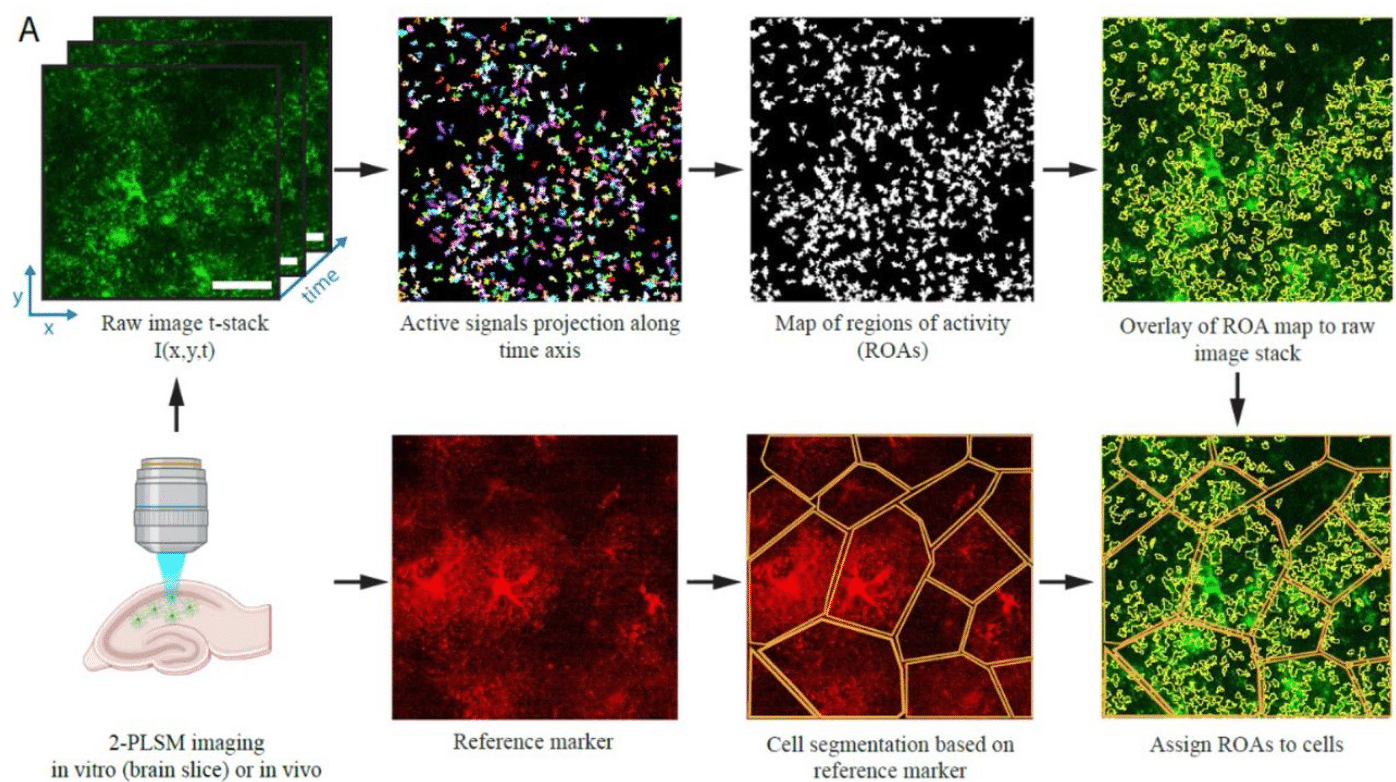
STARDUST: a pipeline for the unbiased analysis of astrocyte regional calcium dynamics
Yifan Wu, Yanchao Dai, Katheryn B. Lefton, Timothy E. Holy, Thomas Papouin
da_Tracker: Automated workflow for high throughput single cell and single phagosome tracking in infected cells
Jacques Augenstreich, Anushka Poddar, Ashton T. Belew, Najib M El-Sayed, Volker Briken
SynBot: An open-source image analysis software for automated quantification of synapses
Justin T. Savage, Juan Ramirez, W. Christopher Risher, Yizhi Wang, Dolores Irala, Cagla Eroglu
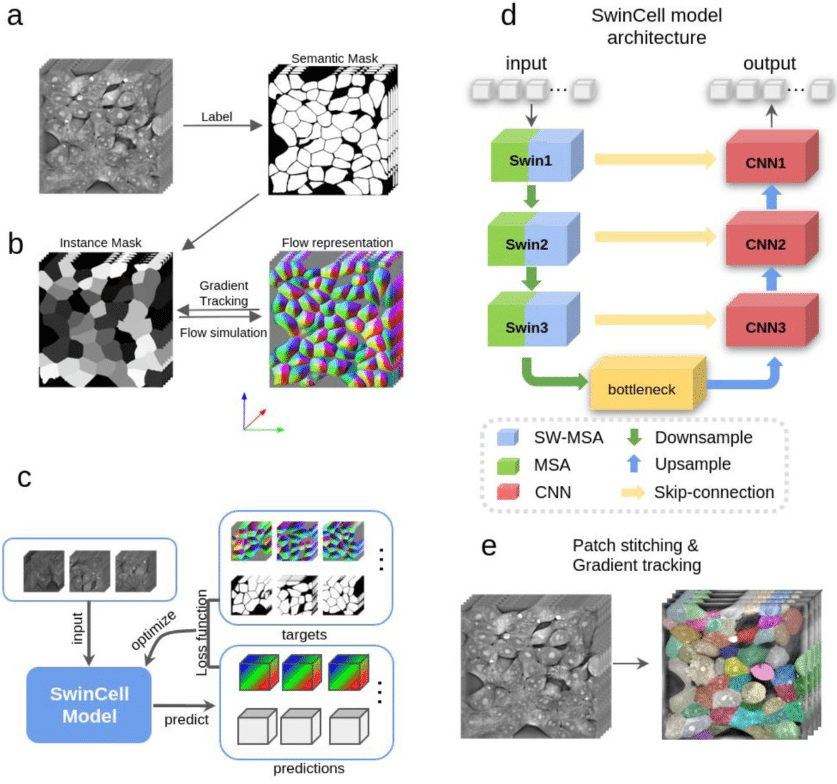
SwinCell: a transformer-based framework for dense 3D cellular segmentation
Xiao Zhang, Zihan Lin, Liguo Wang, Yong S. Chu, Yang Yang, Xianghui Xiao, Yuewei Lin, Qun Liu
OrgaMeas: A pipeline that integrates all the processes of organelle image analysis
Taiki Baba, Akimi Inoue, Susumu Tanimura, Kohsuke Takeda
Self-supervised Vision Transformers for image-to-image labeling: a BiaPy solution to the LightMyCells Challenge
Daniel Franco-Barranco, Aitor González-Marfil, Ignacio Arganda-Carreras
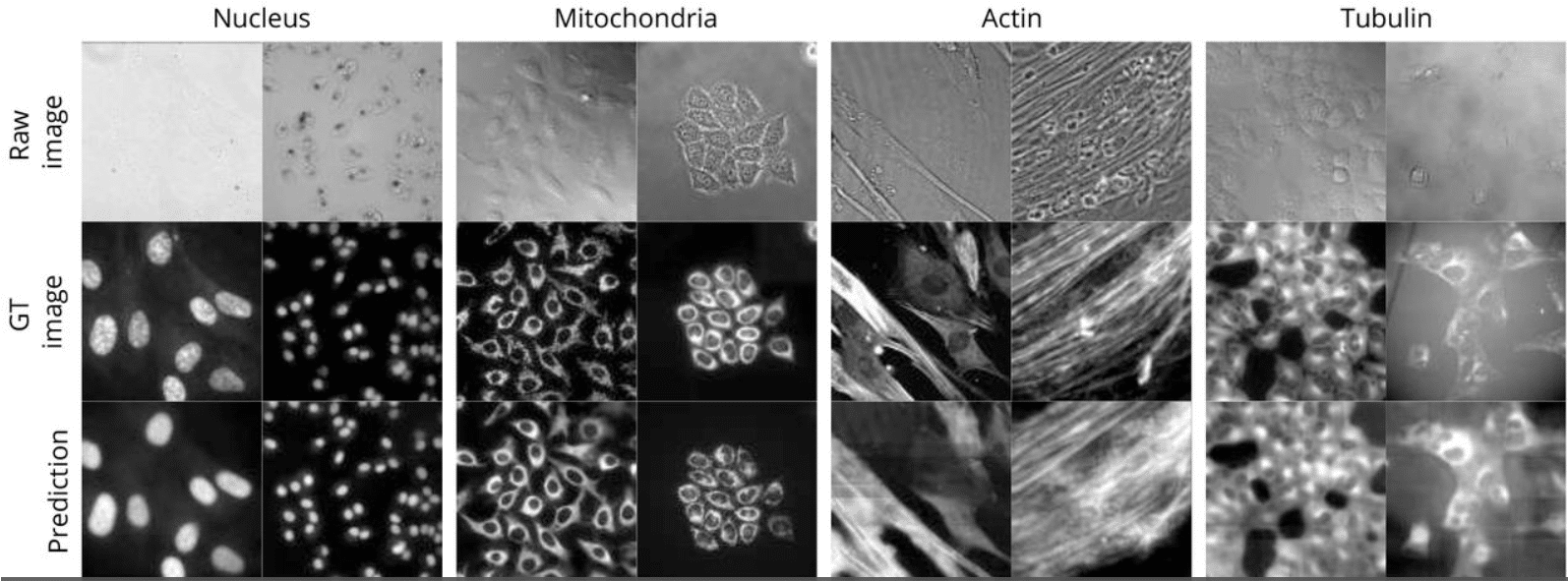
VISION — an open-source software for automated multi-dimensional image analysis of cellular biophysics
Florian Weber, Sofiia Iskrak, Franziska Ragaller, Jan Schlegel, Birgit Plochberger, Erdinc Sezgin, Luca A. Andronico
ViM-UNet: Vision Mamba for Biomedical Segmentation
Anwai Archit, Constantin Pape
Deep learning-driven imaging of cell division and cell growth across an entire eukaryotic life cycle
Shreya Ramakanth, Taylor Kennedy, Berk Yalcinkaya, Sandhya Neupane, Nika Tadic, Nicolas E. Buchler, Orlando Argüello-Miranda
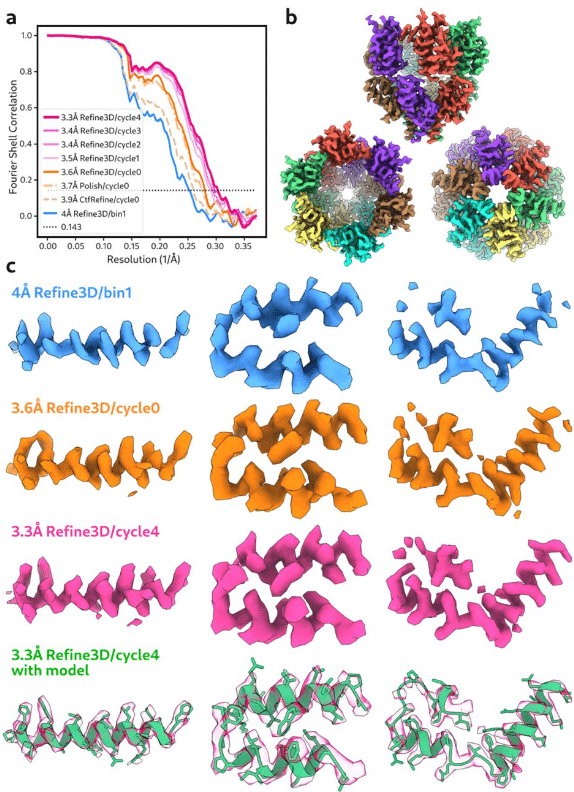
An image processing pipeline for electron cryo-tomography in RELION-5
Alister Burt, Bogdan Toader, Rangana Warshamanage, Andriko von Kügelgen, Euan Pyle, Jasenko Zivanov, Dari Kimanius, Tanmay A.M. Bharat, Sjors H.W. Scheres
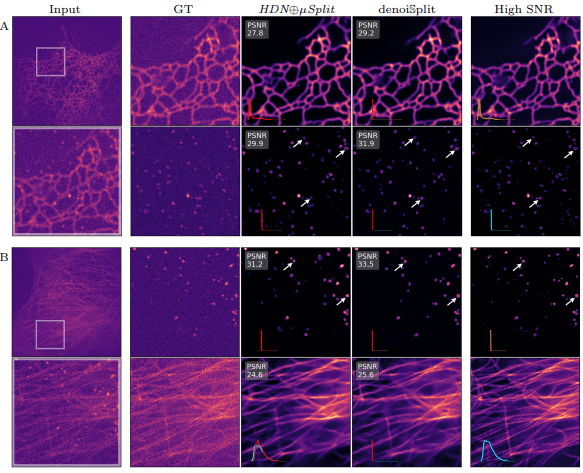
denoiSplit: a method for joint image splitting and unsupervised denoising
Ashesh Ashesh, Florian Jug
Transformers do not outperform Cellpose
Carsen Stringer, Marius Pachitariu
Ais: streamlining segmentation of cryo-electron tomography datasets
Mart G.F. Last, Leoni Abendstein, Lenard M. Voortman, Thomas H. Sharp


 (No Ratings Yet)
(No Ratings Yet)