My history with GFP (and other members of the family)
Posted by Joachim Goedhart, on 7 July 2020
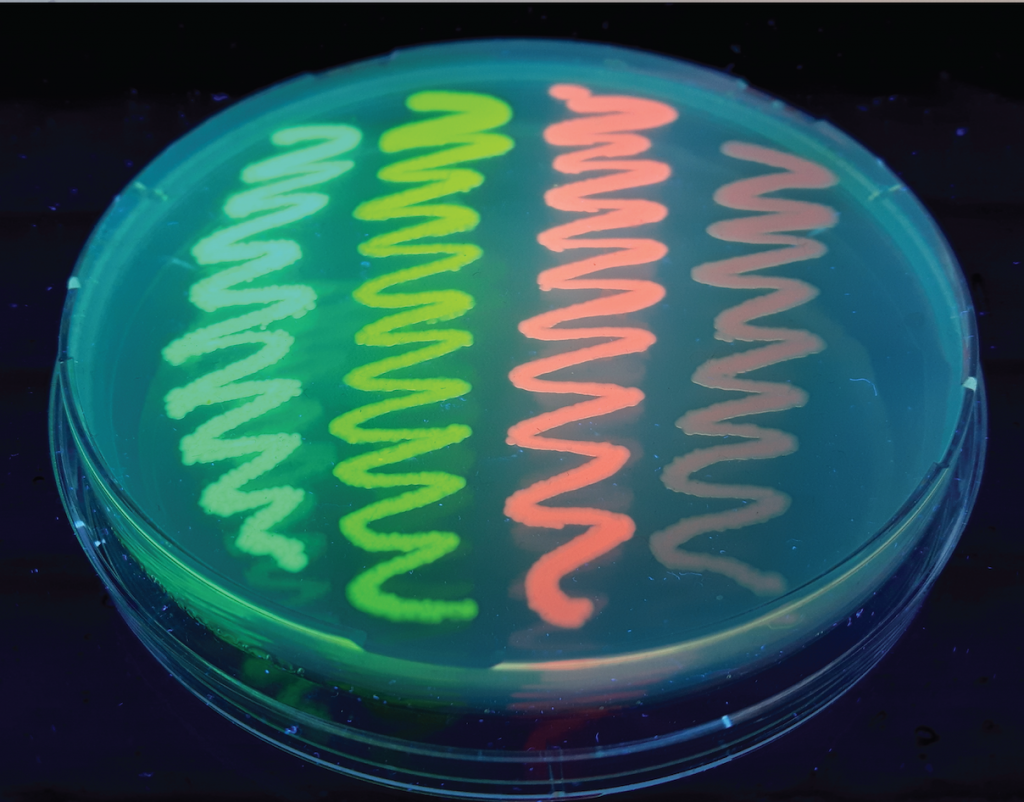
Image made by Eike Mahlandt (University of Amsterdam).
I first crossed paths with green fluorescent protein (GFP) while doing an internship in 1996 at the Max Planck Institute in Göttingen. I was chemically labeling epidermal growth factor (EGF) with the fluorescent dyes Cy3 and Cy5 to study growth-factor signaling. Two post-docs were discussing the use of GFP as an alternative fluorescent tag and my first thought was ‘this will never work’. My reasoning was that the GFP was way too big to be of any use. Today, it is easy to see that I was plain wrong.
Shortly after the internship, I started a PhD project to study a sugary lipid that is an essential signaling molecule for establishing a symbiosis between plants and nitrogen-fixing bacteria. The lipids were labeled with small organic dyes and GFP was only a distant and faint light. A major breakthrough came about when our research building was locked down because of asbestos fibers that were released after drilling holes for cables. To keep going, we visited collaborators in Amsterdam who had experience in molecular biology. During this mini sabbatical, we were immersed in cloning procedures, PCR and ligations (thanks to Ana Laxalt). Being exposed to this technology set the stage for the engineering of fluorescent proteins (FPs) later on.
Although GFP can only be used to tag proteins, it was simultaneously realized by several groups [1,2,3] that protein domains that bind lipids are useful for generating biosensors for lipids. Because of our expertise in fluorescence microscopy, we were contacted by Marten Postma, Jeroen Roelofs and Peter van Haastert who wanted to study lipid signaling in Dictyostelium. We were able to visualize the synthesis of 3-phosphorylayted inositides in Dictyostelium cells with a GFP-based lipid biosensor [4]. This collaborative project was very inspiring and it convinced me of the immense potential of GFP as a probe for fluorescence imaging. I still use a movie that was acquired in those days for teaching.
After my PhD, I moved from Wageningen (agricultural university) to the University of Amsterdam to join the section of Molecular Cytology headed by Dorus Gadella (who was also my thesis advisor). Gert-Jan Kremers joined the group as a PhD candidate and his project was aimed at improving FPs. The mutations that were introduced in the folding- optimized yellow FP Venus were systematically introduced into other FPs to generate a series of FPs that performed well in bacteria: SYFP2, SCFP3A, SBFP2 and SGFP2 [5,6]. The SCFP3A was one of the brightest cyan FPs at that time, but the name was poorly chosen from a marketing point of view.
Dmitry Chudakov and Konstantin Lukyanov were key in the development of photoswitchable FPs and red FPs (mKate, tagRFP) [7,8]. We collaborated with them as part of an EU-funded project, and they explained over dinner at a progress meeting how they screened DNA libraries by using bacterial colonies on plates. With this new insight, we decided to apply our expertise in fluorescence lifetime imaging microscopy to FP screening. Since the excited-state lifetime is (only) related to the quantum yield, it occurred to us that lifetime-based screening could be an elegant way to identify FPs with high quantum yield. Our approach turned out to be highly effective, and it led to the development of mTurquoise [9], a bright cyan FP (with a carefully selected name this time). The next step was made by adding structural information with crucial input from Antoine Royant, leading to the bright cyan variant mTurquoise2 [10]. It is also the monomeric FP with the highest quantum yield (93%) reported to date and this can be largely attributed to the lifetime-screening approach.
Although several monomeric red FPs were reported, there was still a lot of room for improvement. But red FPs pose a much greater engineering challenge. One of the key aims is to achieve fast and full maturation into a ‘red chromophore’ and absence of green and blue forms. Next to this, it is key to prevent oligomerization, which may occur during the screening process, since it increases the brightness of red FPs. Therefore, Dorus Gadella decided to start from a synthetic, consensus red FP template with the strict requirement that it should be monomeric. I was involved in ordering several of the templates as synthetic genes and all of those were failures. At the end of 2011, two PhD candidates (Lindsay Haarbosch and Daphne Bindels) joined the team. They started from mRed7, the first synthetic gene that showed little red fluorescence. Many years later, using a multiparameter screening approach [11] they established the mScarlet series, which represent the brightest monomeric red FPs currently available [12].
I have been lucky enough to witness the maturation of a new series of red FPs while I was busy generating tags and biosensors based on mTurquoise(2). Since mTurquoise2 behaves well in bacteria and mammalian cells, I thought it would work well in any system. I was wrong again. A study by Erik Snapp and coworkers showed that the cellular environment affects FP brightness and reported improved variants for oxidative environments [13]. This study inspired Nils Meiresonne to replace cysteine residues in mTurquoise2 to generate a variant (sfTq2ox) that was optimized for tagging proteins in the periplasm of bacteria [14]. Around the same time, a collaboration with Dennis Botman and Bas Teusink led to identification of a set of optimal FPs for yeast [15]. Similar studies have been performed in other biological systems (plants, worms) to find FPs with optimal properties [16,17]. Together, these studies support the notion that it is important to characterize FP properties in the systems that they are applied to.
Recap and recommendations
Nowadays, there is a wealth of fluorescent proteins to choose from. The FPbase.org is a very useful up-to-date source for finding out about the properties of FPs. For a large majority of FPs, the DNA encoding them can be obtained from addgene.org. The best alternative is to have the gene synthesized by a company and this can be done with the codon usage that you prefer.
But which fluorescent protein do you choose? This challenge was tackled in a highly cited paper by Nathan Shaner and Roger Tsien [18]. Today, there’s even more choice, but also more data, which may help to narrow down the list. I can only give some rough guidelines from my own perspective here. But you can reach out for specific advice (see also this tweet and the responses). In my experience, it is essential to experimentally test some of the key properties of FPs in your own system, since it is a unique combination of equipment, sample and fluorescent probe. Depending on the research question, you may need to examine brightness, oligomerization, photostability, photoconversion [19], pH sensitivity, maturation time [20] or extent, cytotoxicity [21] or some other property in your specific (biological) system. And although FPs have several advantages, it is worth looking at alternatives.
To conclude, I hope to have many more discussions, also on this platform, about fluorescent proteins and probes in general and I encourage everybody to share opinions and experiences.

Joachim Goedhart
Principal Investigator,
Faculty of Science
Swammerdam Institute for Life Sciences
University of Amsterdam


 (2 votes, average: 1.00 out of 1)
(2 votes, average: 1.00 out of 1)