The story behind ZeroCostDL4Mic, or How to get started with using Deep Learning for your microscopy data
Posted by Romain Laine, on 18 January 2021
We all hear again and again about Deep Learning (DL), how great it is, how it’s going to revolutionise science, engineering, medicine and work. But where and how is this all happening? If you wonder how you could get started on thinking, let alone using DL on your microscopy data, look no further, we have the tool for you: ZeroCostDL4Mic [1]. And, like us, you may even get addicted.
It all starts with hitting the wall
As part of the research carried out in the Henriques lab, we develop algorithms for bioimage analysis, especially for the ImageJ/Fiji ecosystem [2]. But for a couple of years, we have been paying close attention to the field of DL applied to microscopy, especially since the publication of the CARE paper [3] highlighting the great potentials of DL to remove noise from fluorescence microscopy data.
So we immediately wondered: How can we use DL on our microscopy data? It looks simple at first: Open the GitHub repository and set up the environment as described. But we hit the wall that many first users of DL encounter: What environment is optimal? What hardware do I need? How do I ensure that all dependencies, such as CUDA and TensorFlow, are compatible and correctly shaking hands? We are not Computer Vision researchers, but it was eventually possible with patience and some dedicated hardware.
But here’s the catch: What if tomorrow you want to run a different DL method based on a different set of libraries and packages? Most likely, you’ll need to set everything up again, from scratch. And what if you don’t even have the required hardware for this?
But, aren’t there lots of cloud-computing platforms to do this? We found out that, yes, several approaches are already taking advantage of these [4]–[6]. Still, they’re difficult to set up and can become costly or cumbersome when you only want to give it a try and merely see if DL is even the right tool for you.
So this was the state of the Henriques lab in late 2018. This was also when we started paying attention to Google Colab and started discussing with cell biologist and image analysis-aficionado Guillaume Jacquemet from Åbo Akademi University, Turku, Finland. Google Colab is an environment within which one can write and execute Python code in a format called notebooks. It then allows us to run that code on cloud-computing provided by Google. Python is the natural language for DL and several libraries/backends are now available for that, including Keras [7] and PyTorch [8].
Notably, within Colab, all the resources necessary for DL are already available (RAM, disk space and good GPUs) and free! Like all good academics, we saw an opportunity to get free stuff and have some fun.
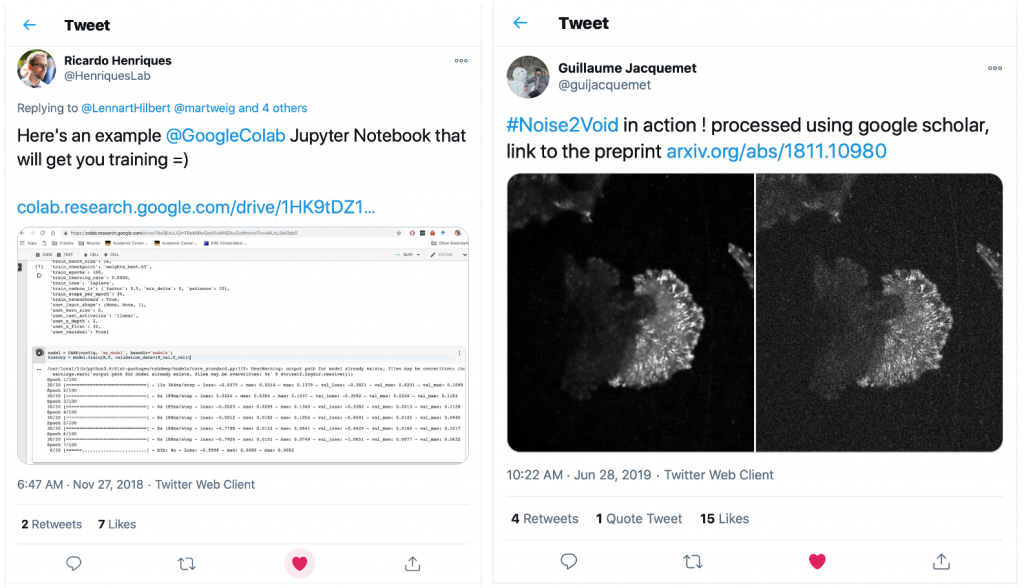
Guillaume had quickly picked up on the capabilities of Google Colab and had put together a Colab version of the fantastic DL-based denoising method Noise2Void [9]. So we put together a team to work on building a set of notebooks for our favourite DL approaches. This is when Lucas von Chamier from the Henriques lab and Johanna Jukkala from the Jacquemet lab joined the adventure.
A need for a general and accessible platform for Deep Learning
At this time, several exciting DL bioimaging applications for e.g. denoising, segmentation or classification had been published and like many others in the bio-imaging community we were excited to try these on our own data. This was also when we realised that many other labs had hit the same wall as us about how to get started with DL and how to test these methods quickly.
That’s when we decided to build a complete platform that would make DL accessible to all, using the freely available resources from Colab.
So, we made a quick checklist:
- Some Python coding skills to port the code from Computer Scientist’s GitHub repositories: Check
- Some basic understanding of DL: Check (thanks to Francois Chollet and his fantastic book [10])
- Data to test our methods on: Check
We first implemented the networks that we were the most excited about and wanted to test on our own images. Common and powerful tasks from DL had been demonstrated for denoising (e.g. CARE [3], Noise2Void [9]), segmentation (U-Net [4], StarDist [11], [12]), predictions of labels from unlabelled data (Label-free prediction – fnet [13]), so we started with those.
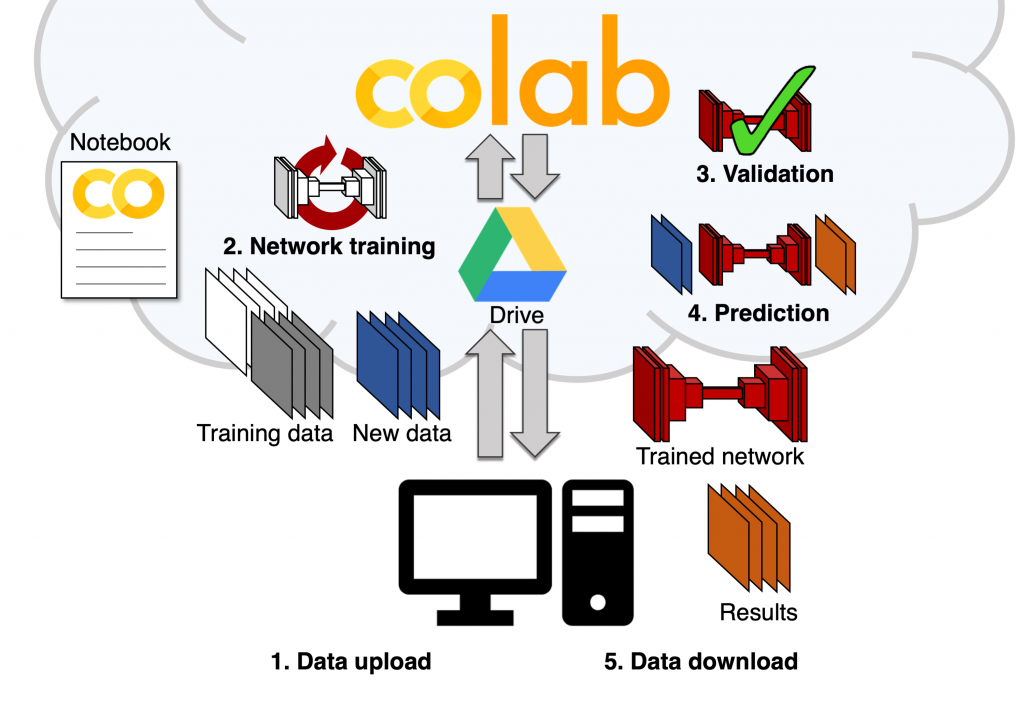
Around that core set of image analysis tasks and neural network types, we built an easy-to-use and templated set of notebooks that includes all the essential steps of a DL workflow: loading the training data (stored on Google Drive), setting some aside for validation, performing data augmentation for diversifying the data even further, completing the training of a new model using the training dataset and then, using the trained model to perform predictions on unknown data (obtaining the denoised image from a new noisy image for instance). We were happy to see that, with the help of the original developers, particularly the teams from Florian Jug and Loic Royer, we could replicate training and predictions on our own data, seamlessly and completely for free. All of this with a simple web-based user interface and no coding necessary for the user.
But here comes a fundamentally important question: How do we know that the trained model’s output is not gibberish?
We need some validation of the performance (or lack thereof) from the models we generate with our platform. Although it is well understood that the output of DL approaches must be carefully evaluated, there are hardly any user-friendly tools for this. In our opinion, the best way to assess the quality of predictions is to quantitatively compare them to ground truth data (the image that we would like the network to output for us in the best case scenario). So we built these approaches into the notebooks themselves in order to ensure we have an easy way to perform evaluation. This gives us some confidence that a particular model provides a good quality output, allowing us to optimise the model for a specific dataset.
Back to the checklist, let’s add an item:
- Make sure it’s useful to others in the field: … We need some Beta testers.
We reached out to colleagues that have their own interests in (and reservations about) DL. And here Seamus Holden’s, Pieta Matila’s and Mike Heileman’s teams came onboard. Especially our favourite microbiologist and beta-tester extraordinaire: Christoph Spahn.
With the essential help and feedback from these teams, we improved the useability of the notebooks and our code to the point where we felt like the project was ready to meet the rest of the world. Our first bioRxiv version of the ZeroCostDL4Mic project was born (March 2020).
With quite a lot of harassment on Twitter (thank you for bearing with this, Twitter-verse), we introduced it to the world. And to our amazement, we received an impressive reception from the community at large and saw adopters cropping up rather quickly. It was absolutely great to see that so many teams had been waiting to get their hands on DL and that our tool could potentially help there. But we needed to keep up with debugging and feeding the beast we had created now.
Feeding the Deep Learning beast
We now needed to get a sense of what was possible and what was not possible with our platform. It was essential to understand if we would hit another wall now. So we decided to get new networks implemented with different approaches and different challenges. We added the capability to do super-resolution microscopy (Deep-STORM [14], thanks to the support of the Shechtman lab), 3D segmentation (3D U-Net [15], thanks to the support from Daniel Krentzel and Martin Jones from the Francis Crick Institute, London) as well as some recent funky GANs (generative adversarial networks) approaches like pix2pix [16] and CycleGAN [17]. These latter ones are peculiar species to apply to microscopy because they were developed for natural scenes and are prone to hallucination relatively easily. But for instance, we were happily surprised to see that pix2pix was able to predict a fake (yes, that’s the dedicated word in the field) DAPI image from an actin label in live-cell fluorescence microscopy data.
Also, motivated by the exciting developments in self-driving cars technologies, we also implemented a fast object detection network called YOLOv2 (standing for You Only Look Once, [18]) and we showed that it was capable of identifying cells of various shapes in live-cell bright-field cell migration assays.
But feeding the DL beast, especially in the framework of Google Colab, meant to check whether we were limited by the amount of data that could be used in Colab (simply due to the disk space and limited time available for each Colab session). We then optimised what could be achieved with the data storage available, notably by making sure that data augmentation and transfer learning were available, and showed that it was possible (a little to our own surprise too) to obtain very high-performance models for all the networks we had implemented.
And here’s a few demonstrations of what we could do with the ZeroCostDL4Mic platform:
Deep Learning for microscopy: a growing community
Now, we are grateful to be part of the ever-growing community of users/developers/enthusiasts of DL for microscopy. We interact closely with teams with a common interest in democratising DL, such as DeepImageJ [19] (notably thanks to Estibaliz Gómez de Mariscal) so that some of our models can be used directly in Fiji, and Wei Ouyang who initiated the great bioimage.io site which we can populate with our notebooks and trained models to easily share with the community.
This is just the beginning of the story and the community surely will sieve through the good, the bad and the ugly [20] that will come from DL, but importantly, we first need to put it in the hands of those who need it the most and who will benefit the most from DL: the biomedical research community. And after all this was our vision for ZeroCostDL4Mic: that our tool help address fundamental biological questions, while building an understanding of its capabilities and limitations.
 | Romain F. Laine MRC-LMCB, University College London Francis Crick Institute, London @LaineBioImaging |


 (3 votes, average: 1.00 out of 1)
(3 votes, average: 1.00 out of 1)