Advances in Expansion Microscopy
Posted by Victoria Alonso, on 4 August 2023
Expansion microscopy was introduced in 2015 by Boyden and his team, revolutionizing the way we see biological samples under a microscope1. Taking advantage of the power of physical enlargement, ExM pushes spatial resolution beyond the diffraction limit, allowing us to observe intricate details at previously unimaginable levels. The underlying principle of ExM lies in a clever combination of chemistry and physics. First, fluorescently labeled molecules are crosslinked to a hydrogel, which serves as a scaffold for the subsequent process. Then, the hydrogel is expanded isotropically with water, causing the sample to enlarge in all three dimensions. This isotropic expansion effectively separates the signals and enables higher-resolution imaging. Some of the most common ExM workflows are summarized in Figure 1 (taken from Zhuang Y. and Shi X. 20232).
This method does not rely on optical or algorithmic improvements and can increase resolution by up to 4.5 times. By embedding fixed biological specimens in a hydrogel matrix and expanding them, structures as small as 70 nm can be resolved using conventional fluorescence microscopy. The expanded samples are optically clear, enabling imaging of thicker specimens that were previously difficult to visualize. The advantages of ExM are significant and were nicely described in Shoh Asano and Ruixuan Gao`s FocalPlane post from 2020 but a lot has happened since then!
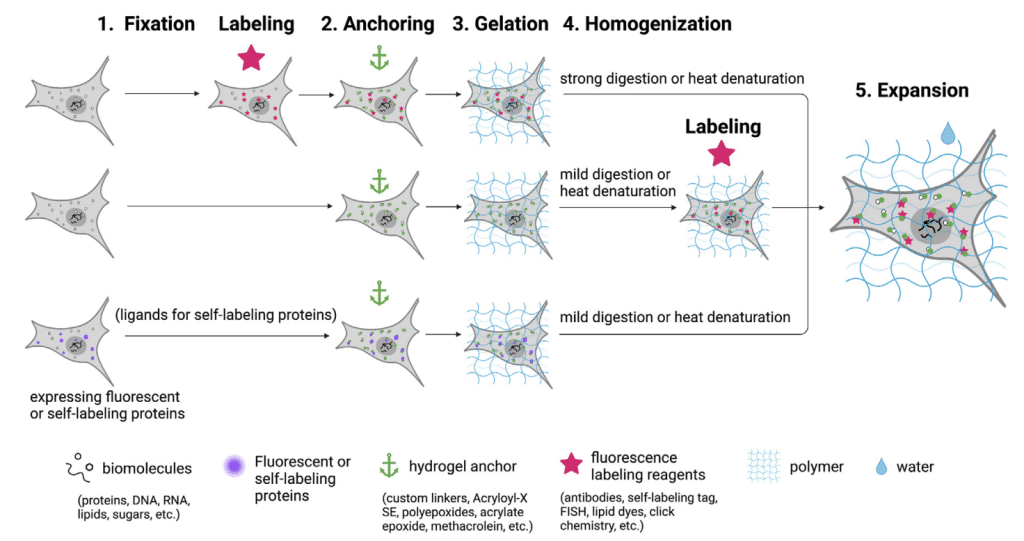
Alternative labeling, gelation, and fixation protocols
An important milestone that facilitated the adoption of ExM in diverse biological fields was the adaptation of immunofluorescence labeling using conventional antibodies3. Nevertheless, its incompatibility with live sample imaging and the reduced signal due to the dispersed fluorophores in the larger sample size is a limitation. To circumvent this problem, many alternatives have arisen in the last few years. For example, label-retention expansion microscopy (LR-ExM) prevents signal loss and greatly enhances labeling efficiency using trifunctional anchors. This technique can be applied to immunofluorescence labeling and also to tagged proteins4. Another alternative is to use fluorescently labeled Fab fragment secondary antibodies to enhance the desired signal5. The use of stabilizer-containing organic fluorophores to mitigate the degradation of dyes due to free radicals has also been explored6. Another approach reported was repeatedly labeling target proteins with a pair of secondary antibodies that specifically bind to each other thus enhancing immunofluorescence signal7. As mentioned, fluorescent proteins can be used for ExM but they are often destroyed during sample preparation. To solve this problem, a method based on a DNA oligonucleotide linked with a fluorescent dye, an acrylamide group (linker), and benzoyl guanine has been reported8. Additionally, new chemically stable Red Fluorescent Proteins have been developed and applied in combination with ExM improving the labeling efficiency9.
Finally, it is very exciting that labeling other biomolecules for ExM is possible. Using click-labeling (Click-ExM) with biotin for example, and then staining with streptavidin conjugated to a fluorophore it is possible to track lipids, glycans, proteins, DNA, RNA, and small molecules10. Lipid expansion microscopy (LExM) has been developed to label phospholipids in cells and organelles membranes11. Other interesting applications that label chromatin and epigenetic marks have been described, for example, ChromExM allows the visualization of chromatin, transcription, and transcription factors in vivo to study nuclear organization at nanoscale12. It is also possible to generate spatial information between epigenetic readers and histone modifications (ExEpi)13.
One of the most remarkable features of ExM is its modular nature, offering flexibility to adapt to various biological samples. In the last couple of years, many alternative gelation protocols have been developed, for example, using vacuum stable gels that allow highly multiplexed staining and are also suitable for coupling with mass-spectrometry assays in tissues14. Another interesting alternative is photopolymerized hydrogels (PhotoExM), which allow for a faster processing time and maintain the integrity of the cells even in 3D cultures15. Also, the use of tetra-gel, a hydrogel that does not depend on free-radical chain-growth polymerization like commonly used polyacrylamide, has been proven useful to improve super-resolution imaging. Tetra-gel diminishes the labeling-placement distortions sometimes observed with polyacrylamide gels16. Finally, a cutting-edge technique called thermo-responsive reversible ExM (T-RevExM) allows the expansion factor to be thermally adjusted in a reversible manner according to the user requirements17.
Another step in the ExM protocol that can be tuned is the fixation approach which in some cases improves the ultrastructural preservation of the sample. For example, the use of cryofixation instead of chemical fixation has allowed the visualization of native cellular components and enhances the availability of epitopes, the downside is that it requires specific equipment18.
Making your samples even bigger and improving the resolution
Recently higher expansion factors have been reached, like Ten-fold Robust Expansion Microscopy (TREx) which allows 10-fold expansion with no specialized equipment of procedures19 or Magnify, which uses a mechanically sturdy gel that retains nucleic acids, proteins, and lipids without the need for a separate anchoring step obtaining an 11-fold expansion20.
Other approaches have relied on combining ExM with different microscopy techniques like light sheet microscopy (ExLLSM) to reconstruct complex 3D structures reducing imaging time, for example in neural circuits21. Also, an innovative technique called “Molecule Anchorable Gel-enabled Nanoscale Imaging of Fluorescence and Stimulated Raman Scattering Microscopy” (MAGNIFIERS) combines stimulated Raman scattering microscopy with ExM, enabling label-free nanoscale imaging of biological targets like proteins, lipids, and DNA22.
In the pursuit of even greater resolution, researchers have combined ExM with super-resolution microscopy techniques. So far, ExM has been used in combination with structured illumination microscopy (ExSIM)23, with stimulated emission depletion microscopy (ExSTED)24 and single-molecule localization microscopy (Ex-SMLM and Ex-dSTORM)25,26. This powerful synergy has pushed resolutions below 20 nm, revealing intricate details at the nanoscale level. More recent applications include the combination of 10-fold expansion microscopy with super-resolution radial fluctuations (SRRF), this new technique is called one-nanometer expansion (ONE) microscopy and allows the visualization of biomolecules at resolutions approaching 1 nm27. ExSRRF has also been applied to pathology specimens28. Furthermore, Klimas and colleagues have combined Magnify with super-resolution optical fluctuation imaging (SOFI) (preLights post from Nadja Hümpfer) and were able to clearly resolve fine details of cilia and basal bodies in human stem-cell-derived lung organoids, observing details previously visualized only with electron microscopy20.
Imaging whole organisms and organs
Expansion microscopy is set to play a crucial role in visualizing and tracing large cellular morphologies and molecular components throughout entire organisms or organs. Recent developments in this direction include the adaptation of ExM to use in 3D organoid models that are difficult to image because they are hard to penetrate with conventional microscopes. Using phototransfer by allyl sulfide exchange-expansion microscopy (PhASE-ExM), Blatchley and colleagues were able to obtain clear samples and perform super-resolution imaging of organoids and their extracellular matrix in 3D29.
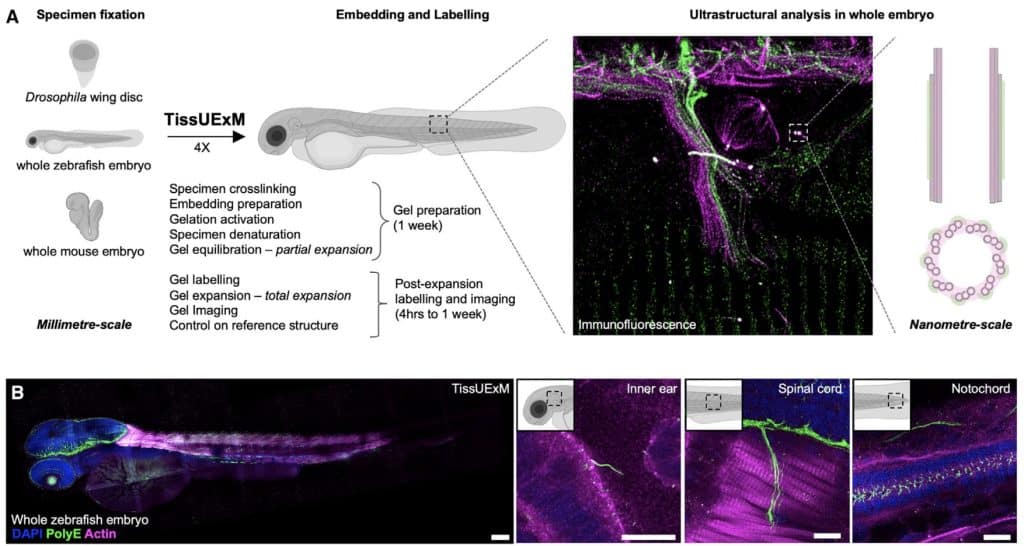
Adaptations of ExM have allowed us to image whole organisms, including ExCel which allows the expansion of C. elegans achieving ~25 nm resolution30, and TissUExM, a method to image whole vertebrate embryos that enables in situ mapping of protein complexes in different organelles31 (Figure 2).
Recently ExM has expanded to plants and fungi, which have the additional complexity of the cell wall, a rigid structure resistant to conventional ExM denaturation and digestion processes32,33. Microorganisms like some types of unicellular fungi and protozoa are sometimes difficult to image due to their small size and in the last couple of years, several ExM protocols have been reported to capture ultrastructural details in these organisms34–38. Especially for protozoan parasites, like Trypanosoma cruzi the causative agent of Chagas disease, I have found ExM very useful to study the particularities of its cytoskeletal network and its single mitochondria39. Very exciting progress has also been made in Plasmodium where the biogenesis and protein distribution of a variety of organelles and structures has been determined using ExM40 (preLights post from Nadja Hümpfer).
Additionally, a recent preprint describes µMagnify, a method derived from ExM that allows imaging of pathogens and infected tissues using a universal biomolecular anchor. This method has a lot of potential for studying how microbes interact with their host systems and will be a powerful tool to develop new diagnosis and treatment strategies against infectious diseases41. It also could be very useful to study host-microbiota interactions.
What´s next?
For centuries, optical microscopy has been a cornerstone in histopathology and microbiology. ExM and its modifications have been tested extensively in different tissue and microbiological samples42. The idea that enlarging the sample could lead to enhanced diagnostic precision is very attractive, especially if we considered the prospect of ExM being adapted to a high-throughput format43, accompanied by the advancement of image processing algorithms and computer power. Another interesting application that has implications for both biomedicine and material sciences is the adaptation of ExM to study cell-material interfaces, for example to study osseointegration of implants44.
Finally, it is interesting to mention a new technique based on the ExM principle, called Unclearing Microscopy (preLights post from Robert Mahen), which allows expanding cells and tissues up to 8000-fold45. This method labeles proteins using NHS-ester ligands linked to biotin in a non-selective manner. Then, they are subsequently exposed to streptavidin bound to horseradish peroxidase and a chromogenic substrate (like 3,3’-diaminobenzidine (DAB) or silver). The samples are then ready to be visualized with the naked eye and if imaged with transmitted light microscopes it is possible to observe details previously only accessible with super-resolution fluorescence or electron microscopy methods.
In conclusion, Expansion Microscopy has opened new frontiers in the world of microscopy, transcending the limits of conventional imaging techniques. Also, it democratizes access to super-resolution microscopy because even labs with standard equipment can set up an ExM workflow that adjusts to their biological sample and needs.
Acronyms for the different methods
| Acronym | Name | Improves |
| ExM | expansion microscopy | The original |
| LR-ExM | label-retention expansion microscopy | labelling |
| Click-ExM | click labelling with expansion microscopy | labelling |
| LExM | lipid expansion microscopy | labelling |
| ChromExM | chromatin expansion microscopy | labelling |
| ExEpi | Expansion microscopy for Epigenetics | labelling |
| PhotoExM | Photo-expansion microscopy | prep time and gel integrity |
| T-RevExM | thermo-responsive reversible ExM | modify expansion factor |
| Cryo-ExM | cryofixation with expansion microscopy | fixation |
| TREx | Ten-fold Robust Expansion Microscopy | increased expansion |
| Magnify | Magnify | increased expansion |
| ExLLSM | Expansion lattice light sheet microscopy | resolution |
| MAGNIFIERS | Molecule Anchorable Gel-enabled Nanoscale Imaging of Fluorescence and Stimulated Raman Scattering Microscopy | resolution |
| ExSIM | expansion with structured illumination microscopy | resolution |
| ExSTED | expansion with stimulated emission depletion microscopy | resolution |
| Ex-SMLM | expansion with single molecular localisation microscopy | resolution |
| Ex-dSTORM | expansion with direct stochastic optical reconstruction microscopy | resolution |
| ExSRRF | Expansion-enhanced super-resolution radial fluctuations | resolution |
| ONE | one-nanometer expansion (ONE) microscopy | resolution |
| Magnify plus SOFI | Magnify with super-resolution optical fluctuation imaging | resolution |
| PhASE-ExM | phototransfer by allyl sulfide exchange-expansion microscopy | extends to new type of sample |
| ExCel | Expansion microscopy with C. elegans | extends to new type of sample |
| TissUExM | tissue ultrastructe expansion microscopy | extends to new type of sample |
| PlantExM | Expansion microscopy of plant cells | extends to new type of sample |
| U-ExM | Ultrastructure expansion microscopy | extends to new type of sample |
| µMagnify | a multiplexed expansion microscopy method for pathogens and infected tissues | extends to new type of sample |
| HiExM | high-throughput expansion microscopy | throughput |
| Unclearing microscopy | Unclearing microscopy | labelling and increased expansion |
References
- Chen, F., Tillberg, P. W. & Boyden, E. S. Expansion Microscopy. Science 347, 543 (2015).
- Zhuang, Y. & Shi, X. Expansion microscopy: A chemical approach for super-resolution microscopy. Curr Opin Struct Biol 81, 102614 (2023).
- Chozinski, T. J. et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nat Methods 13, 485–488 (2016).
- Park, S., Zhuang, Y. & Shi, X. Label-Retention Expansion Microscopy (LR-ExM) Enables Super-Resolution Imaging and High-Efficiency Labeling. J Vis Exp 2022, (2022).
- Sherry, D. M. & Stiles, M. A. Improved fluorescent signal in expansion microscopy using fluorescent Fab fragment secondary antibodies. MethodsX 9, (2022).
- Wen, G., Leen, V., Jia, Y., Rohand, T. & Hofkens, J. Improved Dye Survival in Expansion Microscopy through Stabilizer-Conjugated Linkers. Chemistry – A European Journal 28, e202202404 (2022).
- Yeon, H., Cho, Y., Seo, J., Sim, Y. & Chang, J. B. Simultaneous amplification of multiple immunofluorescence signals via cyclic staining of target molecules using mutually cross-adsorbed antibodies. Scientific Reports 2022 12:1 12, 1–10 (2022).
- Yao, L. et al. Application of SNAP-Tag in Expansion Super-Resolution Microscopy Using DNA Oligostrands. Front Chem 9, 640519 (2021).
- Subach, O. M. et al. LSSmScarlet2 and LSSmScarlet3, Chemically Stable Genetically Encoded Red Fluorescent Proteins with a Large Stokes’ Shift. Int J Mol Sci 23, 11051 (2022).
- Sun, D. en et al. Click-ExM enables expansion microscopy for all biomolecules. Nature Methods 2020 18:1 18, 107–113 (2020).
- M. White, B., Kumar, P., N. Conwell, A., Wu, K. & M. Baskin, J. Lipid Expansion Microscopy. J Am Chem Soc 144, 18212–18217 (2022).
- Pownall, M. E. et al. Chromatin expansion microscopy reveals nanoscale organization of transcription and chromatin. Science (1979) 381, 92–100 (2023).
- Acke, A. et al. Expansion microscopy allows high resolution single cell analysis of epigenetic readers. Nucleic Acids Res 50, (2022).
- Bai, Y. et al. Expanded vacuum-stable gels for multiplexed high-resolution spatial histopathology. Nature Communications 2023 14:1 14, 1–18 (2023).
- Günay, K. A. et al. Photo-expansion microscopy enables super-resolution imaging of cells embedded in 3D hydrogels. Nature Materials 2023 22:6 22, 777–785 (2023).
- Lee, H., Yu, C. C., Boyden, E. S., Zhuang, X. & Kosuri, P. Tetra-gel enables superior accuracy in combined super-resolution imaging and expansion microscopy. Scientific Reports 2021 11:1 11, 1–7 (2021).
- Kang, S. et al. Expansion Microscopy with a Thermally Adjustable Expansion Factor Using Thermoresponsive Biospecimen-Hydrogel Hybrids. ACS Appl Mater Interfaces 13, 28962–28974 (2021).
- Laporte, M. H., Klena, N., Hamel, V. & Guichard, P. Visualizing the native cellular organization by coupling cryofixation with expansion microscopy (Cryo-ExM). Nature Methods 2022 19:2 19, 216–222 (2022).
- Damstra, H. et al. Ten-fold Robust Expansion Microscopy. Bio Protoc 13, (2023).
- Klimas, A. et al. Magnify is a universal molecular anchoring strategy for expansion microscopy. Nature Biotechnology 2023 41:6 41, 858–869 (2023).
- Lillvis, J. L. et al. Rapid reconstruction of neural circuits using tissue expansion and light sheet microscopy. Elife 11, 1–36 (2022).
- Shi, L. et al. Super-Resolution Vibrational Imaging Using Expansion Stimulated Raman Scattering Microscopy. Advanced Science 9, 2200315 (2022).
- Halpern, A. R., Alas, G. C. M., Chozinski, T. J., Paredez, A. R. & Vaughan, J. C. Hybrid Structured Illumination Expansion Microscopy Reveals Microbial Cytoskeleton Organization. ACS Nano 11, 12677–12686 (2017).
- Gao, M., Thielhorn, R., Rentsch, J., Honigmann, A. & Ewers, H. Expansion STED microscopy (ExSTED). Methods Cell Biol 161, 15–31 (2021).
- Kuang, W., Xin, B., Huang, Z. L. & Shi, B. A labeling strategy with effective preservation of fluorophores for expansion single-molecule localization microscopy (Ex-SMLM). Analyst 147, 139–146 (2021).
- Zwettler, F. U., Reinhard, S. & Sauer, M. Ex-dSTORM and automated quantitative image analysis of expanded filamentous structures. Methods Cell Biol 161, 317–340 (2021).
- Shaib, A. H. et al. Visualizing proteins by expansion microscopy. bioRxiv 2022.08.03.502284 (2023) doi:10.1101/2022.08.03.502284.
- Kylies, D. et al. Expansion-enhanced super-resolution radial fluctuations enable nanoscale molecular profiling of pathology specimens. Nature Nanotechnology 2023 18:4 18, 336–342 (2023).
- Blatchley, M. R. et al. In Situ Super-Resolution Imaging of Organoids and Extracellular Matrix Interactions via Phototransfer by Allyl Sulfide Exchange-Expansion Microscopy (PhASE-ExM). Advanced Materials 34, 2109252 (2022).
- Yu, C. C. (Jay), Orozco Cosio, D. M. & Boyden, E. S. ExCel: Super-Resolution Imaging of C. elegans with Expansion Microscopy. Methods Mol Biol 2468, 141–203 (2022).
- Steib, E. et al. TissUExM enables quantitative ultrastructural analysis in whole vertebrate embryos by expansion microscopy. Cell reports methods 2, (2022).
- Hawkins, T. J., Robson, J. L., Cole, B. & Bush, S. J. Expansion Microscopy of Plant Cells (PlantExM). Methods Mol Biol 2604, 127–142 (2023).
- Korovesi, A. G. et al. Expansion Microscopy on Saccharomyces cerevisiae. MicroPubl Biol 2022, (2022).
- Alonso, V. L. Ultrastructure Expansion Microscopy (U-ExM) in Trypanosoma cruzi: localization of tubulin isoforms and isotypes. Parasitol Res 121, (2022).
- Bertiaux, E. et al. Expansion microscopy provides new insights into the cytoskeleton of malaria parasites including the conservation of a conoid. PLoS Biol 19, e3001020 (2021).
- Liffner, B. & Absalon, S. Expansion microscopy reveals plasmodium falciparum blood-stage parasites undergo anaphase with a chromatin bridge in the absence of mini-chromosome maintenance complex binding protein. Microorganisms 9, 2306 (2021).
- Amodeo, S. et al. Characterization of the novel mitochondrial genome segregation factor TAP110 in Trypanosoma brucei. J Cell Sci 134, (2021).
- Dos Santos Pacheco, N. & Soldati-Favre, D. Coupling Auxin-Inducible Degron System with Ultrastructure Expansion Microscopy to Accelerate the Discovery of Gene Function in Toxoplasma gondii. Methods Mol Biol 2369, 121–137 (2021).
- de Hernández, M. A., Peralta, G. M., Vena, R. & Alonso, V. L. Ultrastructural Expansion Microscopy in Three In Vitro Life Cycle Stages of Trypanosoma cruzi. JoVE (Journal of Visualized Experiments) 2023, e65381 (2023).
- Liffner, B. et al. Atlas of Plasmodium falciparum intraerythrocytic development using expansion microscopy. Elife 12, (2023).
- Zhao, Y. et al. MicroMagnify: a multiplexed expansion microscopy method for pathogens and infected tissues. Res Sq (2023).
- Ghosh, S. et al. Expansion microscopy: A revolution in diagnostic pathology. J Microsc 290, 3–9 (2023).
- Day, J. H. et al. HiExM: high-throughput expansion microscopy enables scalable super-resolution imaging. bioRxiv (2023)
- Nakamoto, M. L., Forró, C., Zhang, W., Tsai, C. T. & Cui, B. Expansion Microscopy for Imaging the Cell-Material Interface. ACS Nano (2021).
- M’Saad, O., Shribak, M. & Bewersdorf, J. Unclearing Microscopy. bioRxiv 2022.11.29.518361 (2022).


 (No Ratings Yet)
(No Ratings Yet)