FocalPlane playlist: Rita Strack
Posted by Rita Strack, on 7 February 2024
I was really excited to be invited to create a blog post with my very own “microscopy mix tape” on the FocalPlane blog. I am a huge fan of the work The Company of Biologists does, and I am a consummate consumer of all things FocalPlane as well. Just by way of quick introduction, I am a microscopy geek who happens to also have been the editor handling microscopy, imaging, and probes at Nature Methods for the past ~nine years. I love methods that let us see the invisible, and I truly believe that methods drive biology.
Here, borrowing heavily from Christophe Leterrier’s awesome playlist, I will begin with recent preprints and papers that have caught my eye, and I’ll end by highlighting some great FocalPlane content.
Playlist Part 1: recent papers and preprints
Scientists often ask me what editors think about preprints, and while I certainly can’t speak for all of us, I for one love them! I think they’re an awesome way to rapidly disseminate scientific discovery, a wonderful way to find out about the coolest new work (to invite to the journal!), and I also think they can serve as a landmark to show how papers are improved during the process of peer review. My only complaint is that there are too many good ones to keep up with! Actually, the site formerly known as Twitter helps me a lot with that (you can find me @rita_strack).
So here are some papers that have caught my eye lately, in no particular order of importance, and I hope it goes without saying that this is non-exhaustive list!
- Enabling Global Image Data Sharing in the Life Sciences
Peter Bajcsy, Sreenivas Bhattiprolu, Katy Boerner, Beth A Cimini, Lucy Collinson, Jan Ellenberg, Reto Fiolka, Maryellen Giger, Wojtek Goscinski, Matthew Hartley, Nathan Hotaling, Rick Horwitz, Florian Jug, Anna Kreshuk, Emma Lundberg, Aastha Mathur, Kedar Narayan, Shuichi Onami, Anne L. Plant, Fred Prior, Jason Swedlow, Adam Taylor, Antje Keppler
arXiv 23 Jan 2024 - Harmonizing the Generation and Pre-publication Stewardship of FAIR Image Data
Nikki Bialy, Frank Alber, Brenda Andrews, Michael Angelo, Brian Beliveau, Lacramioara Bintu, Alistair Boettiger, Ulrike Boehm, Claire M. Brown, Mahmoud Bukar Maina, James J. Chambers, Beth A. Cimini, Kevin Eliceiri, Rachel Errington, Orestis Faklaris, Nathalie Gaudreault, Ronald N. Germain, Wojtek Goscinski, David Grunwald, Michael Halter, Dorit Hanein, John W. Hickey, Judith Lacoste, Alex Laude, Emma Lundberg, Jian Ma, Leonel Malacrida, Josh Moore, Glyn Nelson, Elizabeth Kathleen Neumann, Roland Nitschke, Shuichi Onami, Jaime A. Pimentel, Anne L. Plant, Andrea J. Radtke, Bikash Sabata, Denis Schapiro, Johannes Schöneberg, Jeffrey M. Spraggins, Damir Sudar, Wouter-Michiel Adrien Maria Vierdag, Niels Volkmann, Carolina Wählby, Siyuan (Steven)Wang, Ziv Yaniv, Caterina Strambio-De-Castillia
arXiv 23 Jan 2024 - MIFA: Metadata, Incentives, Formats, and Accessibility guidelines to improve the reuse of AI datasets for bioimage analysis
Teresa Zulueta-Coarasa, Florian Jug, Aastha Mathur, Josh Moore, Arrate Muñoz-Barrutia, Liviu Anita, Kola Babalola, Pete Bankhead, Perrine Gilloteaux, Nodar Gogoberidze, Martin Jones, Gerard J. Kleywegt, Paul Korir, Anna Kreshuk, Aybüke Küpcü Yoldaş, Luca Marconato, Kedar Narayan, Nils Norlin, Bugra Oezdemir, Jessica Riesterer, Norman Rzepka, Ugis Sarkans, Beatriz Serrano, Christian Tischer, Virginie Uhlmann, Vladimír Ulman, Matthew Hartley
arXiv 17 Nov 2023
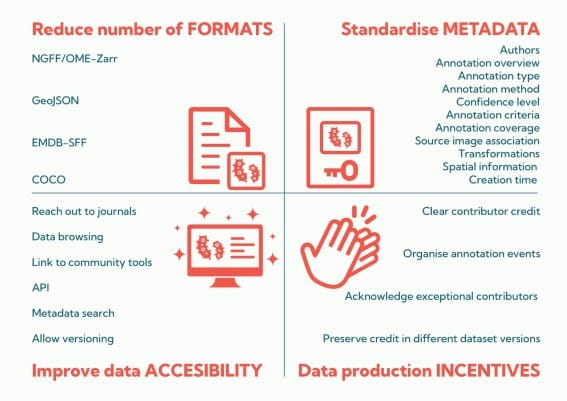
Figure extracted from Zulueta-Coarasa, et al. The image is made available under a CC-BY 4.0 International license.
These three white papers draw attention to the great need for FAIR (findable, accessible, interoperable, and reusable) data sharing in the microscopy community. Although many challenges hinder the broad sharing of microscopy data, including dataset size, data diversity, file formats, expense, metadata reporting, and a dearth of image data repositories, the time is now to address this challenge. Sharing microscopy data is critically important for data reuse and reproducibility, but will also be critical for the future of machine intelligence in bioimaging. Beyond infrastructure, what is needed is community buy-in that microscopy data should be maintained in a way that are mineable beyond the person who generated them and shared broadly for the broader scientific enterprise. In this way, the field is still in its infancy and far behind fields like genomics and structural biology. These white papers lay out guidance for how to bridge the gap, and I hope they get broad community engagement.
- Visual interpretability of image-based classification models by generative latent space disentanglement applied to in vitro fertilization
Oded Rotem, Tamar Schwartz, Ron Maor, Yishay Tauber, Maya Tsarfati Shapiro, Marcos Meseguer, Daniella Gilboa, Daniel S. Seidman, Assaf Zaritsky
bioRxiv 17 Nov 2023
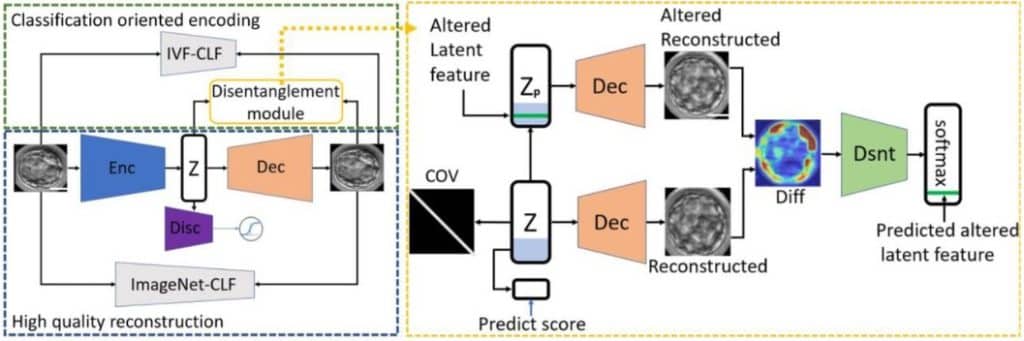
Deep learning has impacted all aspects of bioimaging and bioimage analysis, but the black box nature of these algorithms leaves many skeptics and hold outs in their wake. Therefore, it is critically important that we learn how the models are learning. I really like how this paper untangles what the deep learning classifier “sees” when it chooses how to classify human embryos by quality.
- Projective oblique plane structured illumination microscopy
Bo-Jui Chang, Douglas Shepherd & Reto Fiolka
npj Imaging 1: 2 (2023)
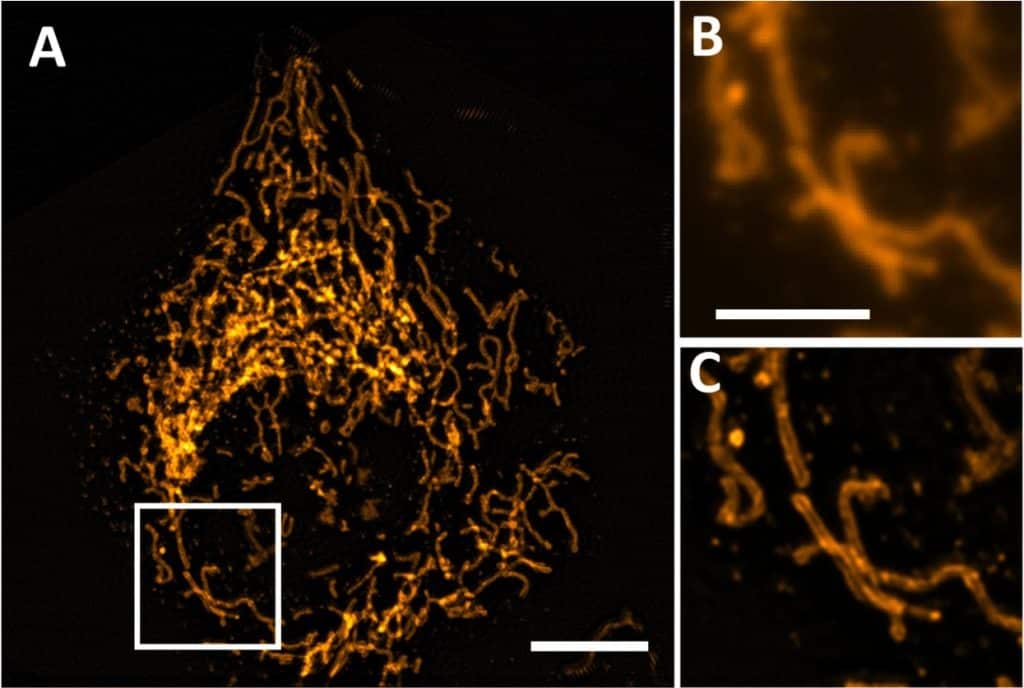
I wanted to highlight this paper for two reasons: first, the science is awesome, as it’s the fastest 3D cellular SIM to date! They achieve that using oblique plane SIM and projection imaging (something Nature Methods knows a little about :D). The second reason is that it’s the first paper ever published by the journal npj Imaging, a journal for which I serve as a consulting editor that is poised to be a really important journal for bio and biomedical imaging across scales.
- Single-molecule detection and super-resolution imaging with a portable and adaptable 3D-printed microscopy platform (Brick-MIC)
Gabriel G. Moya Muñoz, Oliver Brix, Philipp Klocke, Nicolas D. Wendler, Eitan Lerner, Niels Zijstra, Thorben Cordes
bioRxiv 29 Dec 2023
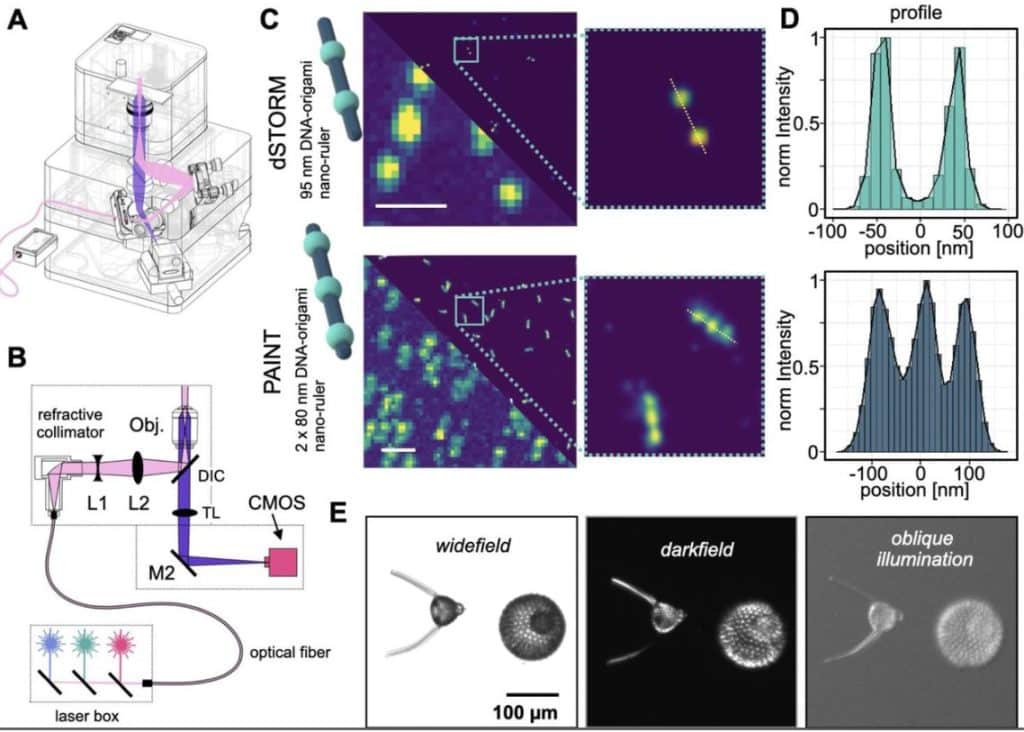
- An open-source, high resolution, automated fluorescence microscope
Ando C. Zehrer, Ana Martin-Villalba, Benedict Diederich, Helge Ewers
bioRxiv 16 Jan 2024
The open microscopy hardware community has really been flourishing in recent years, and researchers across the world now have more access than ever to diverse microscopy modalities. These two recent preprints really embody this trend. In the first, the Cordes lab establishes the Brick-MIC platform for making diverse microscopes. I got to see a few versions of the system first-hand when I visited Gabriel in Munich, and they showed me a single-molecule confocal that used a laser pointer for illumination, and still offered state-of-the-art imaging capabilities.
The second from Diederich and Ewers builds on the well-established UC2 microscopy toolbox, and offers remarkable image quality at a fraction of the cost of comparable commercial microscopes. I think innovations in this space are pivotal for democratizing microscopy and inspiring the next generation of microscopists and engineers
Playlist part 2: great FocalPlane content
Staying on the topic of open microscopy, I’d like to point you readers to this excellent blog post on the OpenFlexure microscope from a few years ago. I got to meet Richard Bowman last year and was really impressed with the work they’ve done building and disseminating this open source system. I think the impact of this project will be felt especially strongly in the Global South, where it could improve both basic research and clinical diagnostics.
I also love the blog series Bio Image Analysis with Napari by Mara Lampert. I am old school in the sense that I grew up writing little plug ins for ImageJ/FIJI and living my best life. That being said, I am so impressed with napari and how quickly it’s made an impact in the bioimaging community. This blog series is so practically useful, it really helps connects users with the technology. I feel that these conversations between developers and biologists are great, especially when they work both ways, so that users can tell developers what they need (or pain points) and expert users and developers can help others unlock the full potential of a tool.
Finally, I’ve been having fun reading the Latin American Microscopists series. As an editor, a big part of my job is networking, and knowing who is doing what and where. It can be hard to keep these efforts up globally, especially when the bulk of my travel has me visiting the US and Europe. Therefore, I am so enthusiastic about learning about scientists outside my network to stay in the know and to help myself build future connections. These are also very enjoyable to read. Honestly, I wish I could listen to these interviews as chats while I do my morning exercise!
Well, if you’ve made it all this way, thanks so much for reading! I hope I’ve raised awareness of some cool content. Cheers!


 (2 votes, average: 1.00 out of 1)
(2 votes, average: 1.00 out of 1)