Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 19 June 2024
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis.
A Postdoctoral Training Program in Bioimage Analysis
Beth A Cimini, Callum Tromans-Coia, David Stirling, Suganya Sivagurunathan, Rebecca Senft, Pearl Ryder, Esteban Miglietta, Paula Llanos, Nasim Jamali, Barbara Diaz-Rohrer, Shatavisha Dasgupta, Mario Cruz, Erin Weisbart, Anne E Carpenter
VASCilia (Vision Analysis StereoCilia): A Napari Plugin for Deep Learning-Based 3D Analysis of Cochlear Hair Cell Stereocilia Bundles
Yasmin M. Kassim, David B. Rosenberg, Alma Renero, Samprita Das, Samia Rahman, Ibraheem Al Shammaa, Samer Salim, Zhuoling Huang, Kevin Huang, Yuzuru Ninoyu, Rick Adam Friedman, Artur A Indzhykulian, Uri Manor
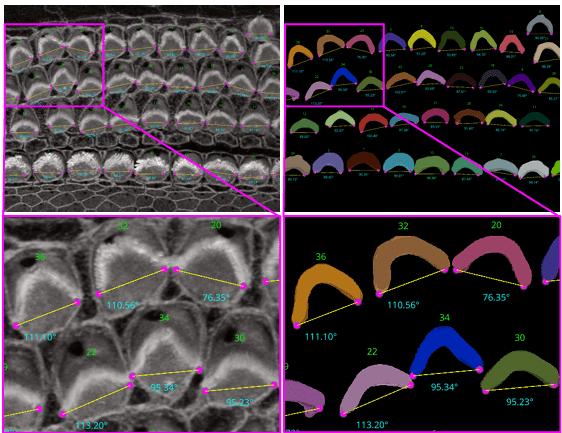
RESPAN: an accurate, unbiased and automated pipeline for analysis of dendritic morphology and dendritic spine mapping
Sergio B. Garcia, Alexa P. Schlotter, Daniela Pereira, Franck Polleux, Luke A. Hammond
A new protocol for multispecies bacterial infections in zebrafish and their monitoring through automated image analysis
Désirée A. Schmitz, Tobias Wechsler, Hongwei Bran Li, Bjoern H. Menze, Rolf Kümmerli
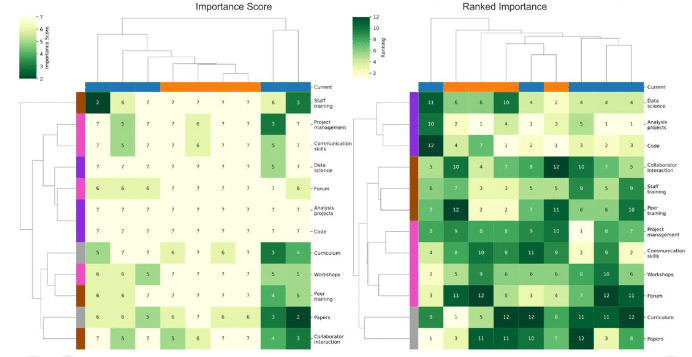
Piximi – An Images to Discovery web tool for bioimages and beyond
Levin M Moser, Nodar Gogoberidze, Andréa Papaleo, Alice Lucas, David Dao, Christoph A Friedrich, Lassi Paavolainen, Csaba Molnar, David R Stirling, Jane Hung, Rex Wang, Callum Tromans-Coia, Bin Li, Edward L Evans III, Kevin W Eliceiri, Peter Horvath, Anne E Carpenter, Beth A Cimini
Figure extracted from Moser, et al.. The image is made available under a CC-BY 4.0 International license.
2D Label-free Prediction of Multiple Organelles Across Different Transmitted-light Microscopy Images with Bag-of-Experts
Yu Zhou, Shuo Zhao, Justin Sonneck, Jianxu Chen
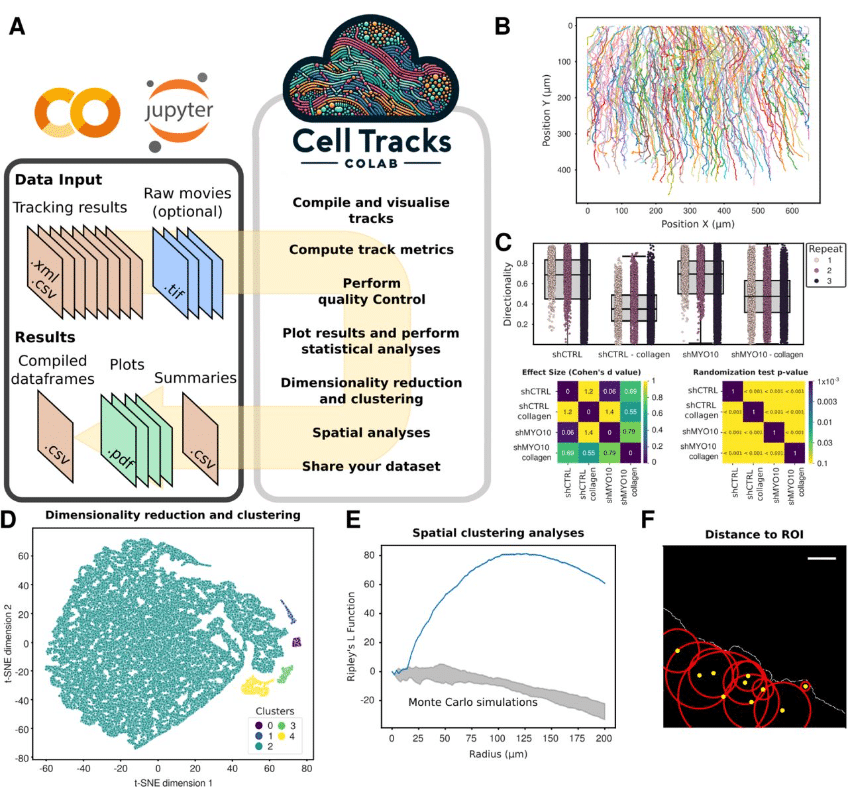
CellTracksColab — A platform for compiling, analyzing, and exploring tracking data
Estibaliz Gómez-de-Mariscal, Hanna Grobe, Joanna W. Pylvänäinen, Laura Xénard, Ricardo Henriques, Jean-Yves Tinevez, Guillaume Jacquemet
Surforama: interactive exploration of volumetric data by leveraging 3D surfaces
Kevin A. Yamauchi, Lorenz Lamm, Lorenzo Gaifas, Ricardo D. Righetto, Daniil Litvinov, Benjamin D. Engel, Kyle Harrington
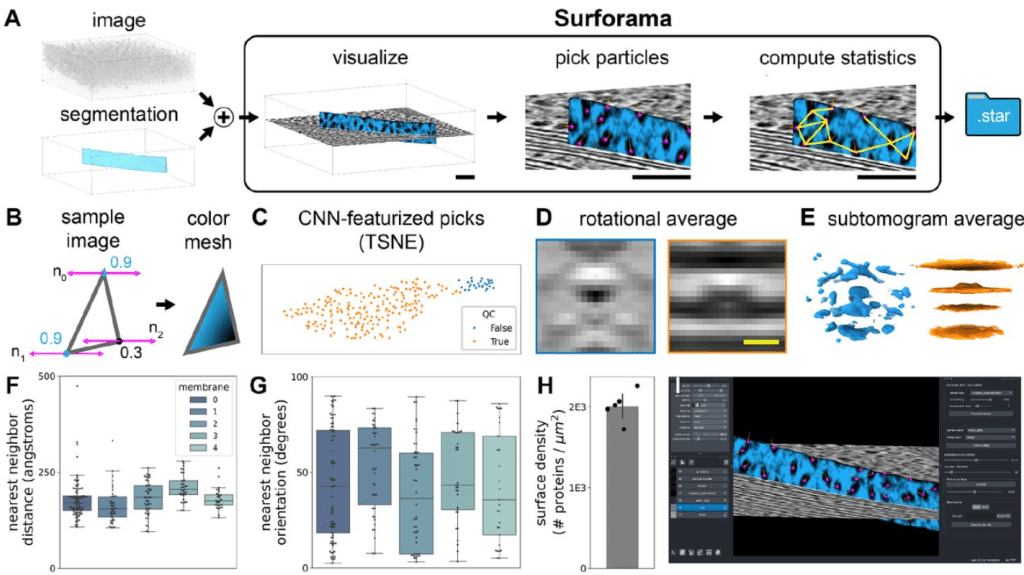
Figure extracted from Yamauchi, et al.. The image is made available under a CC-BY 4.0 International license.
ESPRESSO: Spatiotemporal omics based on organelle phenotyping
Lorenzo Scipioni, Giulia Tedeschi, Mariana Navarro, Yunlong Jia, Scott Atwood, Jennifer A. Prescher, Michelle Digman
Baikal: Unpaired Denoising of Fluorescence Microscopy Images using Diffusion Models
Shivesh Chaudhary, Sivaramakrishnan Sankarapandian, Matt Sooknah, Joy Pai, Caroline McCue, Zhenghao Chen, Jun Xu
Enhanced Cell Tracking Using A GAN-based Super-Resolution Video-to-Video Time-Lapse Microscopy Generative Model
Abolfazl Zargari, Najmeh Mashhadi, S. Ali Shariati
A Self-Supervised Learning Approach for High Throughput and High Content Cell Segmentation
Van Lam, Jeff M. Byers, Michael Robitaille, Logan Kaler, Joseph A. Christodoulides, Marc P. Raphael
OoCount: A Machine-Learning Based Approach to Mouse Ovarian Follicle Counting and Classification
Lillian Folts, Anthony S. Martinez, Corey Bunce, Blanche Capel, Jennifer McKey
Cellpose as a reliable method for single-cell segmentation of autofluorescence microscopy images
Jeremiah M Riendeau, Amani A Gillette, Emmanuel Contreras Guzman, Mario Costa Cruz, Aleksander Kralovec, Shirsa Udgata, Alexa Schmitz, Dustin A Deming, Beth A Cimini, Melissa C Skala
Unsupervised deep learning enables blur-free super-resolution in two-photon microscopy
Haruhiko Morita, Shuto Hayashi, Takahiro Tsuji, Daisuke Kato, Hiroaki Wake, Teppei Shimamura
A scalable and modular computational pipeline for axonal connectomics: automated tracing and assembly of axons across serial sections
Russel Torres, Kevin Takasaki, Olga Gliko, Connor Laughland, Wan-Qing Yu, Emily Turschak, Ayana Hellevik, Pooja Balaram, Eric Perlman, Uygar Sümbül, R. Clay Reid
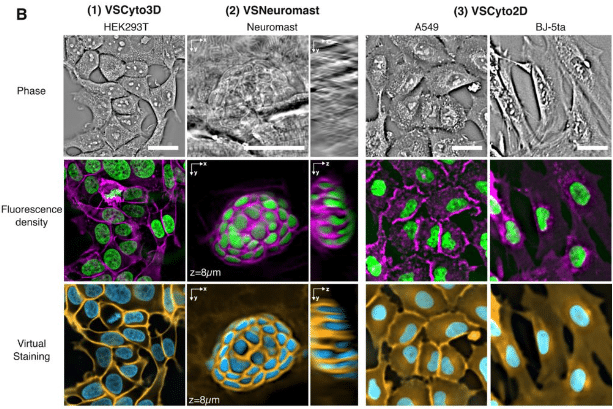
Robust virtual staining of landmark organelles
Ziwen Liu, Eduardo Hirata-Miyasaki, Soorya Pradeep, Johanna Rahm, Christian Foley, Talon Chandler, Ivan Ivanov, Hunter Woosley, Tiger Lao, Akilandeswari Balasubramanian, Chad Liu, Manu Leonetti, Carolina Arias, Adrian Jacobo, Shalin B. Mehta
DeepKymoTracker: A tool for accurate construction of cell lineage trees for highly motile cells
Khelina Fedorchuk, Sarah M. Russell, Kajal Zibaei, Mohammed Yassin, Damien G Hicks
High-Resolution In Silico Painting with Generative Models
Trang Le, Emma Lundberg
Deep learning-based cytoskeleton segmentation for accurate high-throughput measurement of cytoskeleton density
Ryota Horiuchi, Asuka Kamimura, Yuga Hanaki, Hikari Matsumoto, Minako Ueda, Takumi Higaki
EzReverse – a Web Application for Background Adjustment of Color Images
Xinwei Song, Joachim Goedhart
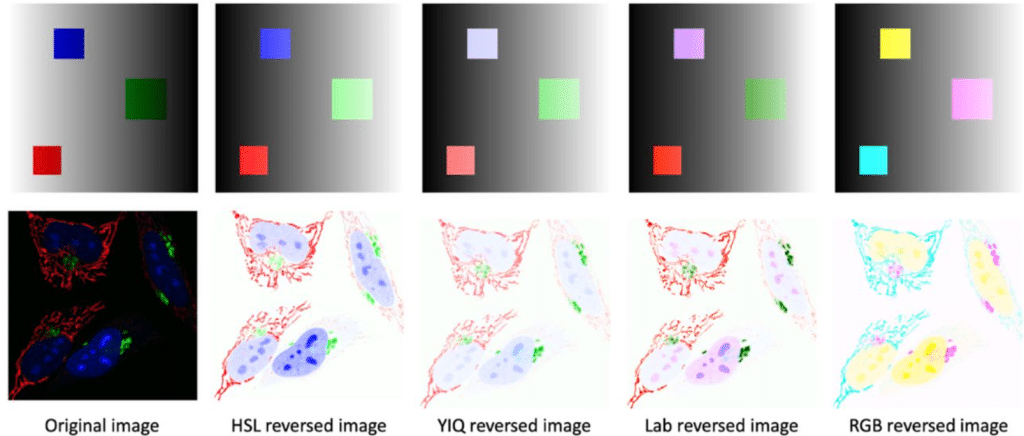
Figure extracted from Song and Goedhart. The image is made available under a CC-BY-NC 4.0 International license.
COEXIST: Coordinated single-cell integration of serial multiplexed tissue images
Robert T. Heussner, Cameron F. Watson, Christopher Z. Eddy, Kunlun Wang, Eric M. Cramer, Allison L. Creason, Gordon B. Mills, Young Hwan Chang
Automated Prediction of Fibroblast Phenotypes Using Mathematical Descriptors of Cellular Features
Alex Khang, Abigail Barmore, Georgios Tseropoulos, Kaustav Bera, Dilara Batan, Kristi S. Anseth
Learning precise segmentation of neurofibrillary tangles from rapid manual point annotations
Sina Ghandian, Liane Albarghouthi, Kiana Nava, Shivam R. Rai Sharma, Lise Minaud, Laurel Beckett, Naomi Saito, Charles DeCarli, Robert A. Rissman, Andrew F. Teich, Lee-Way Jin, Brittany N. Dugger, Michael J. Keiser
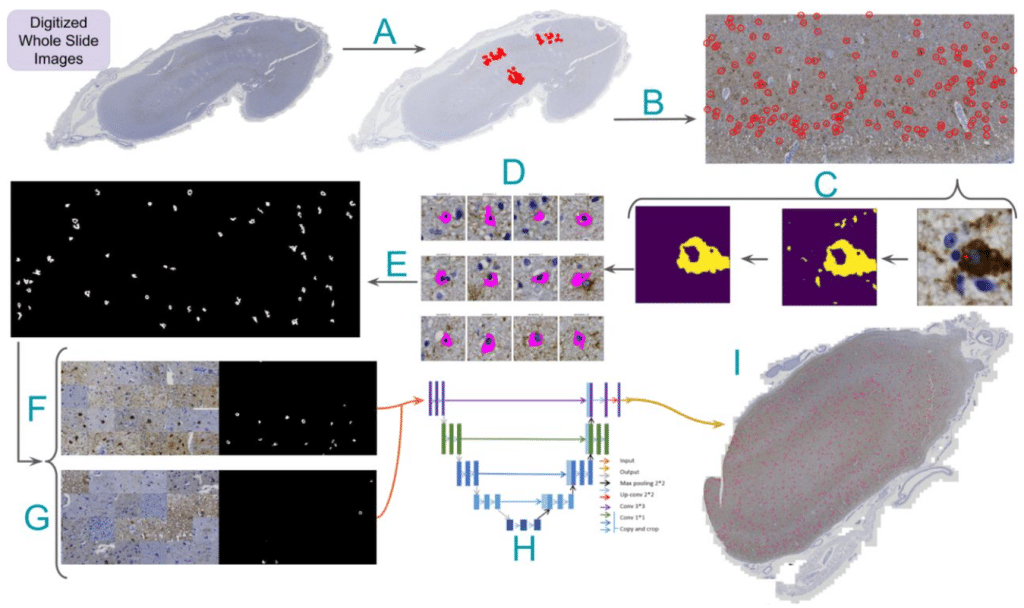
QuST: QuPath Extension for Integrative Whole Slide Image and Spatial Transcriptomics Analysis
Chao-Hui Huang


 (No Ratings Yet)
(No Ratings Yet)