Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 29 July 2024
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
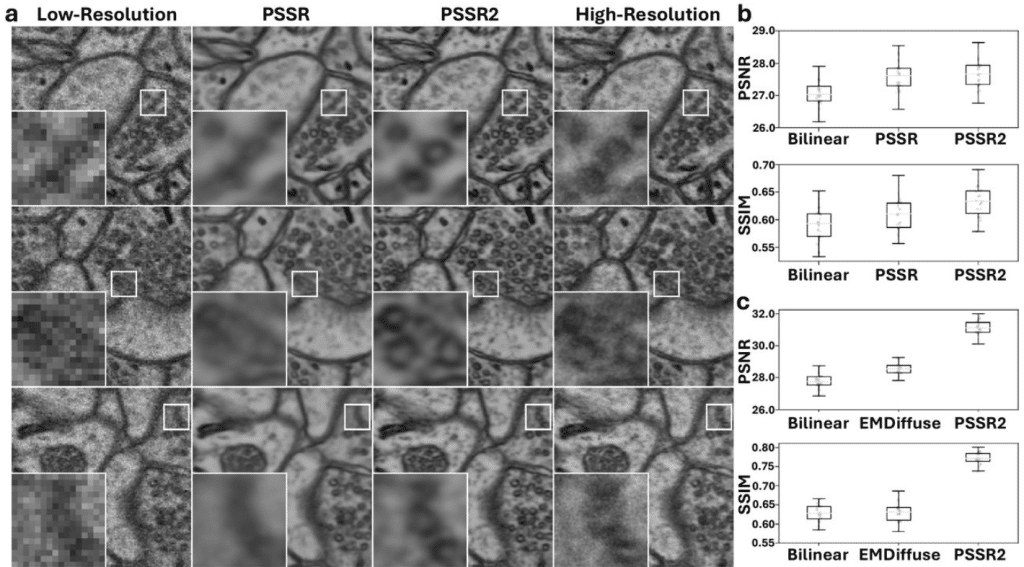
PSSR2: a user-friendly Python package for democratizing deep learning-based point-scanning super-resolution microscopy
Hayden Stites, Uri Manor
Time-lapse Image Super-resolution Neural Network with Reliable Confidence Quantification for Optical Microscopy
Chang Qiao, Shuran Liu, Yuwang Wang, Wencong Xu, Xiaohan Geng, Tao Jiang, Jingyu Zhang, Quan Meng, Hui Qiao, Dong Li, Qionghai Dai
Implicit Neural Image Field for Biological Microscopy Image Compression
Gaole Dai, Cheng-Ching Tseng, Qingpo Wuwu, Rongyu Zhang, Shaokang Wang, Ming Lu, Tiejun Huang, Yu Zhou, Ali Ata Tuz, Matthias Gunzer, Jianxu Chen, Shanghang Zhang
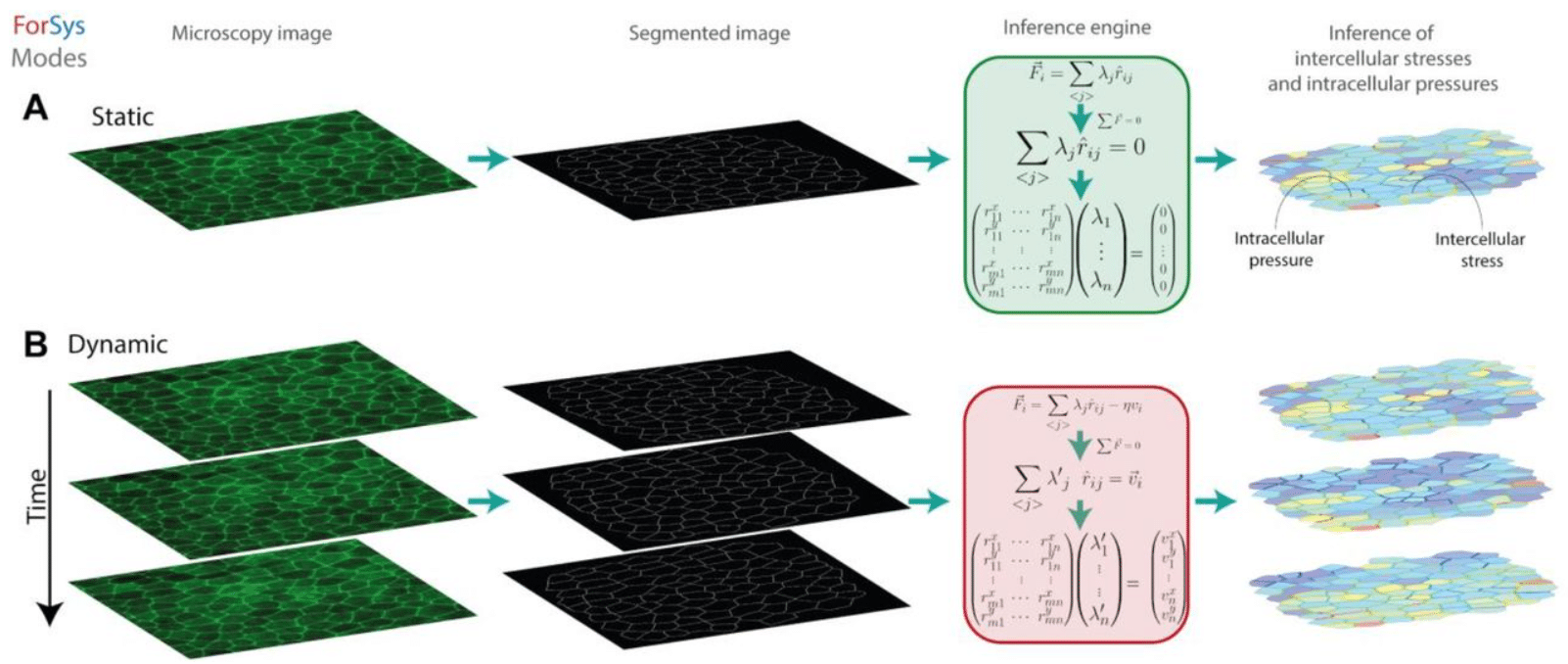
ForSys: non-invasive stress inference from time-lapse microscopy
Augusto Borges, Jerónimo R. Miranda-Rodríguez, Alberto Sebastián Ceccarelli, Guilherme Ventura, Jakub Sedzinski, Hernán López-Schier, Osvaldo Chara
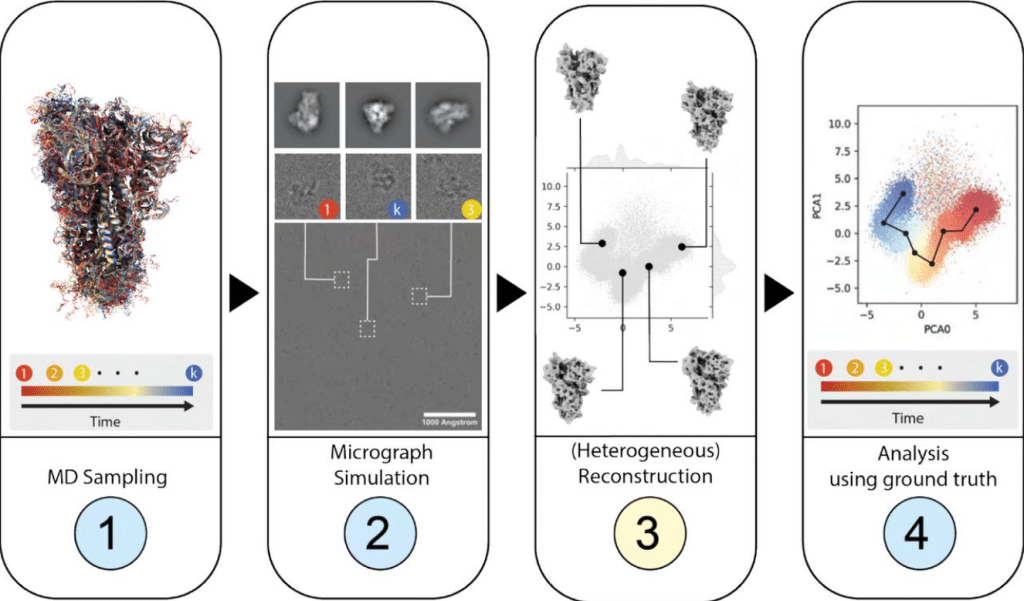
Roodmus: A toolkit for benchmarking heterogeneous electron cryo-microscopy reconstructions
Maarten Joosten, Joel Greer, James Parkhurst, Tom Burnley, Arjen J. Jakobi
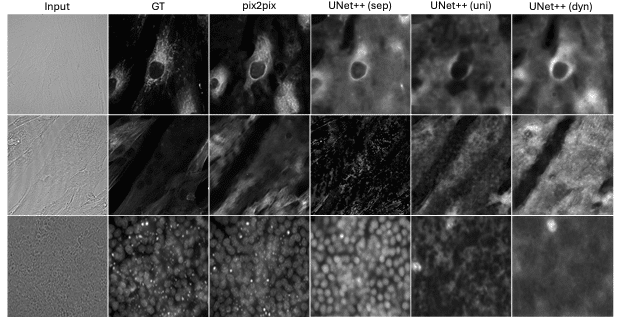
Predicting fluorescent labels in label-free microscopy images with pix2pix and adaptive loss in Light My Cells challenge
Han Liu, Hao Li, Jiacheng Wang, Yubo Fan, Zhoubing Xu, Ipek Oguz
AutoMorFi: Automated Whole-image Morphometry in Fiji/ImageJ for Diverse Image Analysis Needs
Ouzéna Bouadi, Chenkai Yao, Jason Zeng, Danielle Beason, Nyomi Inda, Zoe Malone, Jonathan Yoshihara, Amritha Vinayak Manjally, Clifton Johnson III, Jonathan Cherry, Chin-Yi Chen, Tzu-Chieh Huang, Bogdana Popovic, Maria Henley, Guangmei Liu, Elizabeth Kharitonova, Ella Zeldich, Hannah Aichelman, Sarah W. Davies, Peter Walentek, Yuan Tian, Hengye Man, Elif Ozsen, Kristen Harder, Thomas D. Gilmore, David Pitt, Tuan Leng Tay
Imaging single particle profiler to study nanoscale bioparticles using conventional confocal microscopy
Taras Sych, André Görgens, Loïc Steiner, Gozde Gucluer, Ylva Huge, Farhood Alamdari, Markus Johansson, Firas Aljabery, Amir Sherif, Susanne Gabrielsson, Samir EL Andaloussi, Erdinc Sezgin
CryoSamba: self-supervised deep volumetric denoising for cryo-electron tomography data
Jose Inacio Costa-Filho, Liam Theveny, Marilina de Sautu, Tom Kirchhausen
Setting up an institutional OMERO environment for bioimage data: perspectives from both facility staff and users
Anett Jannasch, Silke Tulok, Chukwuebuka William Okafornta, Thomas Kugel, Michele Bortolomeazzi, Tom Boissonnet, Christian Schmidt, Andy Vogelsang, Claudia Dittfeld, Sems-Malte Tugtekin, Klaus Matschke, Leocadia Paliulis, Dirk Lindemann, Gunar Fabig, Thomas Müller-Reichert
Mew: Multiplexed Immunofluorescence Image Analysis through an Efficient Multiplex Network
Sukwon Yun, Jie Peng, Alexandro E. Trevino, Chanyoung Park, Tianlong Chen
QuST: QuPath Extension for Integrative Whole Slide Image and Spatial Transcriptomics Analysis
Chao-Hui Huang
FRAP analysis Measuring biophysical kinetic parameters using image analysis
Sharva V. Hiremath, Etika Goyal, Gregory T. Reeves, Cranos M. Williams
Cross-correlation image analysis for real-time particle tracking
Leonardo R. Werneck, Cody Jessup, Austin Brandenberger, Tyler Knowles, Charles W. Lewandowski, Megan Nolan, Ken Sible, Zachariah B. Etienne, Brian D’Urso
Benchmarking Large Language Models for Bio-Image Analysis Code Generation
Robert Haase, Christian Tischer, Jean-Karim Hériché, Nico Scherf

Morphological simulation tests the limits on phenotype discovery in 3D image analysis
Rachel A. Roston, Sophie M. Whikehart, Sara M. Rolfe, Murat Maga
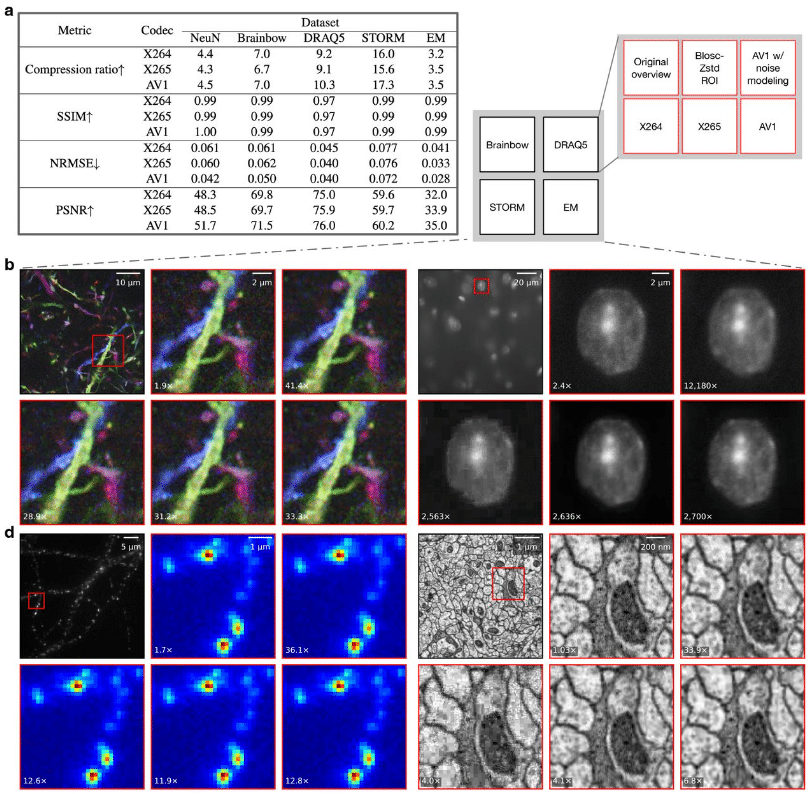
Artifact-Minimized High-Ratio Image Compression with Preserved Analysis Fidelity
Bin Duan, Logan A Walker, Bin Xie, Wei Jie Lee, Alexander Lin, Yan Yan, Dawen Cai
TissueMap: A Web-Based Multiplexed Image Viewer
D.G.P. van IJzendoorn, M. Matusiak, R.B. West, M. van de Rijn
SPACEc: A Streamlined, Interactive Python Workflow for Multiplexed Image Processing and Analysis
Yuqi Tan, Tim N. Kempchen, Martin Becker, Maximilian Haist, Dorien Feyaerts, Yang Xiao, Graham Su, Andrew J. Rech, Rong Fan, John W. Hickey, Garry P. Nolan
Image processing tools for petabyte-scale light sheet microscopy data
Xiongtao Ruan, Matthew Mueller, Gaoxiang Liu, Frederik Görlitz, Tian-Ming Fu, Daniel E. Milkie, Joshua L. Lillvis, Alexander Kuhn, Johnny Gan Chong, Jason Li Hong, Chu Yi Aaron Herr, Wilmene Hercule, Marc Nienhaus, Alison N. Killilea, Eric Betzig, Srigokul Upadhyayula
EM-Compressor: Electron Microscopy Image Compression in Connectomics with Variational Autoencoders
Yicong Li, Core Francisco Park, Daniel Xenes, Caitlyn Bishop, Daniel R. Berger, Aravi D.T. Samuel, Brock Wester, Jeff W. Lichtman, Hanspeter Pfister, Wanhua Li, Yaron Meirovitch
PENGUIN: A rapid and efficient image preprocessing tool for multiplexed spatial proteomics
A. M. Sequeira, M. E. Ijsselsteijn, M. Rocha, Noel F.C.C. de Miranda
From in vitro to in silico: a pipeline for generating virtual tissue simulations from real image data
Elina Nürnberg, Mario Vitacolonna, Roman Bruch, Markus Reischl, Rüdiger Rudolf, Simeon Sauer
MitoStructSeg: A Comprehensive Platform for Mitochondrial Structure Segmentation and Analysis
Xinsheng Wang, Buqing Cai, Zhuo Jia, Yuanbo Chen, Shuai Guo, Zheng Liu, Xiaohua Wan, Fa Zhang, Bin Hu
Mantis: high-throughput 4D imaging and analysis of the molecular and physical architecture of cells
Ivan E. Ivanov, Eduardo Hirata-Miyasaki, Talon Chandler, Rasmi Cheloor-Kovilakam, Ziwen Liu, Soorya Pradeep, Chad Liu, Madhura Bhave, Sudip Khadka, Carolina Arias, Manuel D. Leonetti, Bo Huang, Shalin B. Mehta
SPArrOW: a flexible, interactive and scalable pipeline for spatial transcriptomics analysis
Lotte Pollaris, Bavo Vanneste, Benjamin Rombaut, Arne Defauw, Frank Vernaillen, Julien Mortier, Wout Vanhenden, Liesbet Martens, Tinne Thoné, Jean-Francois Hastir, Anna Bujko, Wouter Saelens, Jean-Christophe Marine, Hilde Nelissen, Evelien Van Hamme, Ruth Seurinck, Charlotte L. Scott, Martin Guilliams, Yvan Saeys
OPUS-TOMO: Deep Learning Framework for Structural Heterogeneity Analysis in Cryo-electron Tomography
Zhenwei Luo, Qinghua Wang, Jianpeng Ma
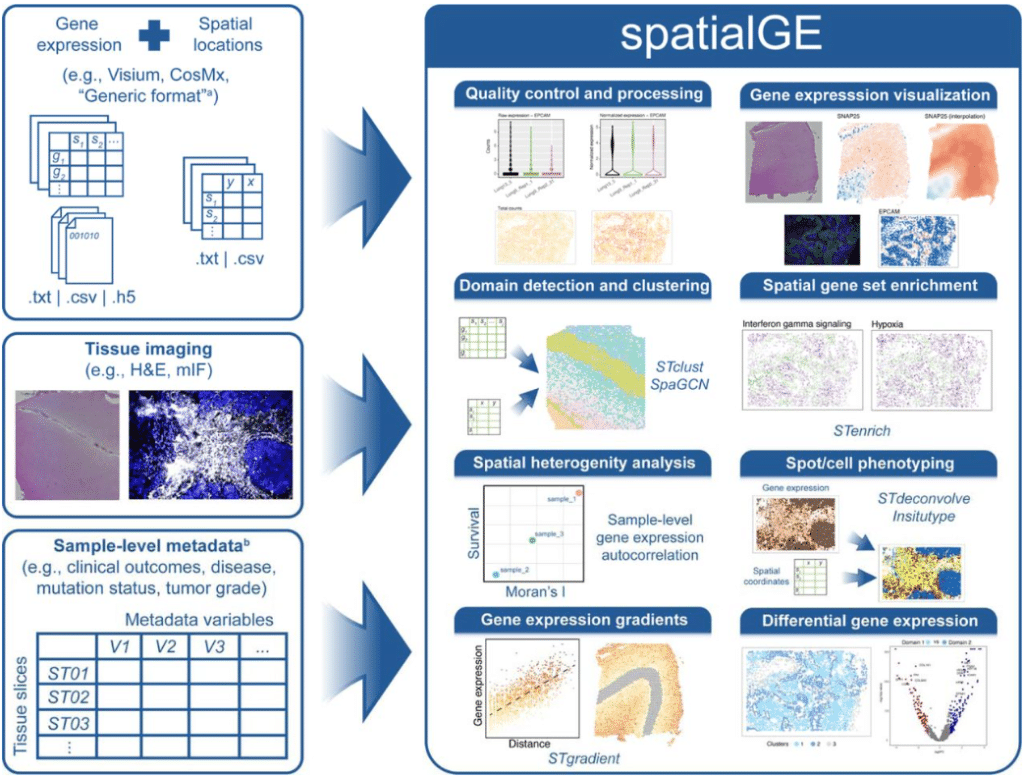
spatialGE: A user-friendly web application to democratize spatial transcriptomics analysis
Oscar E. Ospina, Roberto Manjarres-Betancur, Guillermo Gonzalez-Calderon, Alex C. Soupir, Inna Smalley, Kenneth Tsai, Joseph Markowitz, Ethan Vallebuona, Anders Berglund, Steven Eschrich, Xiaoqing Yu, Brooke L. Fridley


 (No Ratings Yet)
(No Ratings Yet)