Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 6 September 2024
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
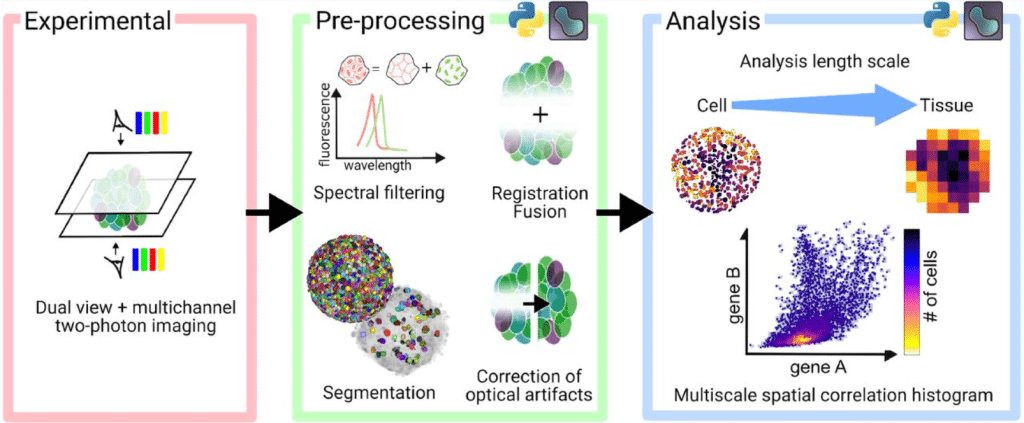
A quantitative pipeline for whole-mount deep imaging and multiscale analysis of gastruloids
Alice Gros, Jules Vanaret, Valentin Dunsing-Eichenauer, Agathe Rostan, Philippe Roudot, Pierre-François Lenne, Léo Guignard, Sham Tlili
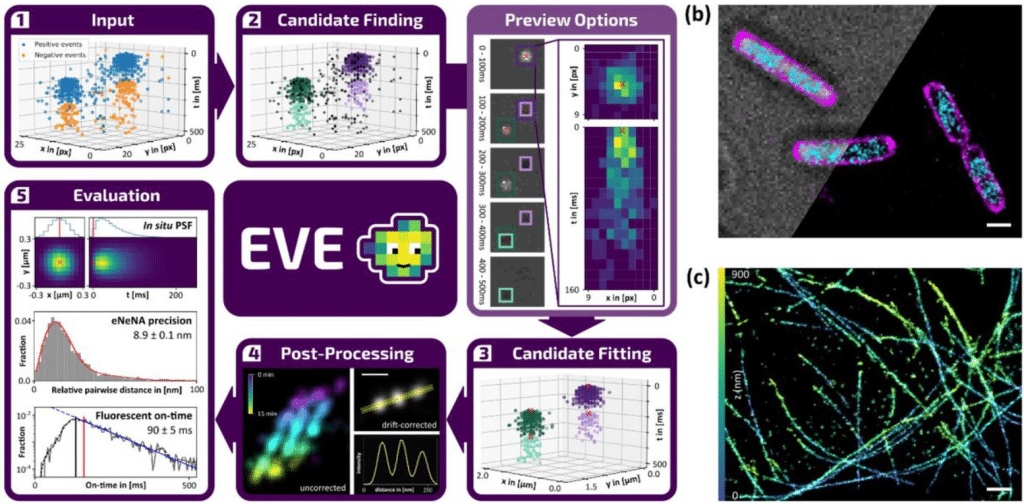
EVE is an open modular data analysis software for event-based localization microscopy
Laura M. Weber, Koen J.A. Martens, Clément Cabriel, Joel J. Gates, Manon Albecq, Fredrik Vermeulen, Katarina Hein, Ignacio Izeddin, Ulrike Endesfelder
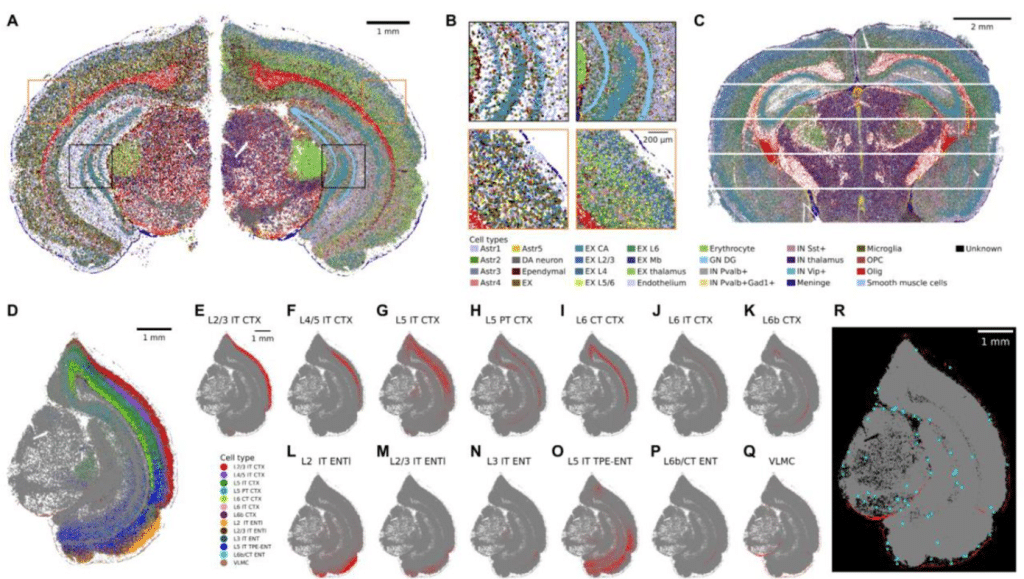
Sainsc: a computational tool for segmentation-free analysis of in-situ capture
Niklas Müller-Bötticher, Sebastian Tiesmeyer, Roland Eils, Naveed Ishaque
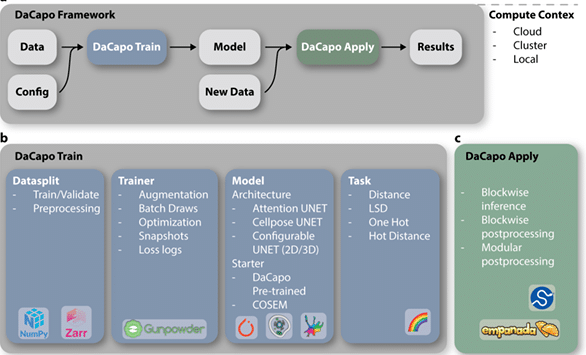
DaCapo: a modular deep learning framework for scalable 3D image segmentation
William Patton, Jeff L. Rhoades, Marwan Zouinkhi, David G. Ackerman, Caroline Malin-Mayor, Diane Adjavon, Larissa Heinrich, Davis Bennett, Yurii Zubov, CellMap Project Team, Aubrey V. Weigel, Jan Funke
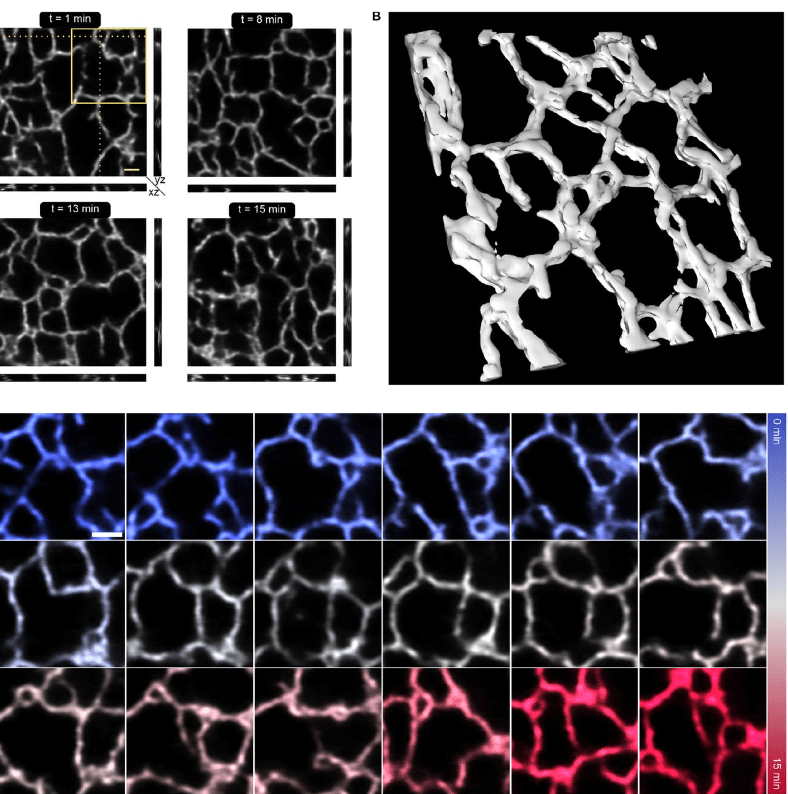
Fast and long-term super-resolution imaging of ER nano-structural dynamics in living cells using a neural network
Johanna V. Rahm, Ashwin Balakrishnan, Maren Wehrheim, Alexandra Kaminer, Marius Glogger, Laurell F. Kessler, Matthias Kaschube, Hans-Dieter Barth, Mike Heilemann
PyPlaque: an Open-source Python Package for Phenotypic Analysis of Virus Plaque Assays
Trina De, Vardan Andriasyan, Artur Yakimovich
Cecelia: a multifunctional image analysis toolbox for decoding spatial cellular interactions and behaviour
Dominik Schienstock, Jyh Liang Hor, Sapna Devi, Scott N. Mueller
FourierMIL: Fourier filtering-based multiple instance learning for whole slide image analysis
Yi Zheng, Harsh Sharma, Margrit Betke, Jennifer E. Beane, Vijaya B. Kolachalama
Novel high-content and open-source image analysis tools for profiling mitochondrial morphology in neurological cell models
Marcus Y. Chin, David A. Joy, Madhuja Samaddar, Anil Rana, Johann Chow, Takashi Miyamoto, Meredith Calvert
CRISP: Correlation-Refined Image Segmentation Process
Jennifer K. Briggs, Erli Jin, Matthew J. Merrins, Richard K.P. Benninger
TissueViewer: A Web-Based Multiplexed Image Viewer
D.G.P. van IJzendoorn, M. Matusiak, R.B. West, M. van de Rijn
Image retrieval based on closed-loop visual–semantic neural decoding
Ryohei Fukuma, Takufumi Yanagisawa, Hidenori Sugano, Kentaro Tamura, Satoru Oshino, Naoki Tani, Yasushi Iimura, Hui Ming Khoo, Hiroharu Suzuki, Huixiang Yang, Takamitsu Iwata, Madoka Nakajima, Shinji Nishimoto, Yukiyasu Kamitani, Haruhiko Kishima
3D-Aligner: An advanced computational tool designed to correct image distortion in expansion microscopy for precise 3D reconstitution and quantitative analysis
Jonathan Loi, Dhaval Ghone, Xiaofei Qu, Aussie Suzuki
Live Cell Painting: image-based profiling in live cells using Acridine Orange
Fernanda Garcia-Fossa, Thaís Moraes-Lacerda, Mariana Rodrigues-da-Silva, Barbara Diaz-Rohrer, Shantanu Singh, Anne E. Carpenter, Beth A. Cimini, Marcelo Bispo de Jesus
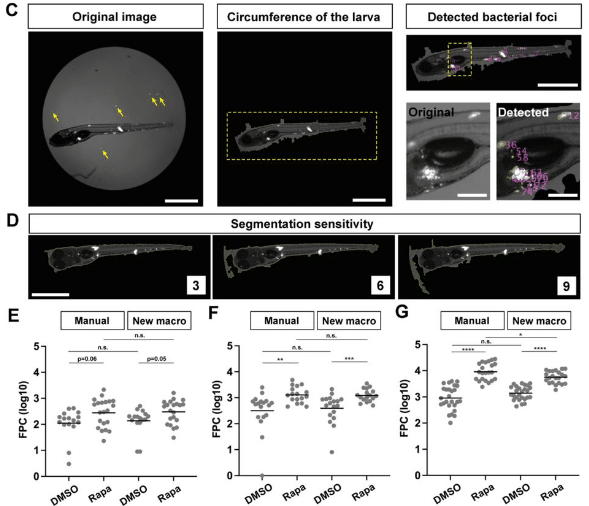
An Image Processing Tool for Automated Quantification of Bacterial Burdens in Zebrafish Larvae
Naoya Yamaguchi, Hideo Otsuna, Michal Eisenberg-Bord, Lalita Ramakrishnan
Generative AI for Cell Type-Specific Fluorescence Image Generation of hPSC-derived Cardiac Organoid
Arun Kumar Reddy Kandula, Tanakit Phamornratanakun, Angello Huerta Gomez, Marcel El-Mokahal, Zhen Ma, Yunhe Feng, Huaxiao Yang
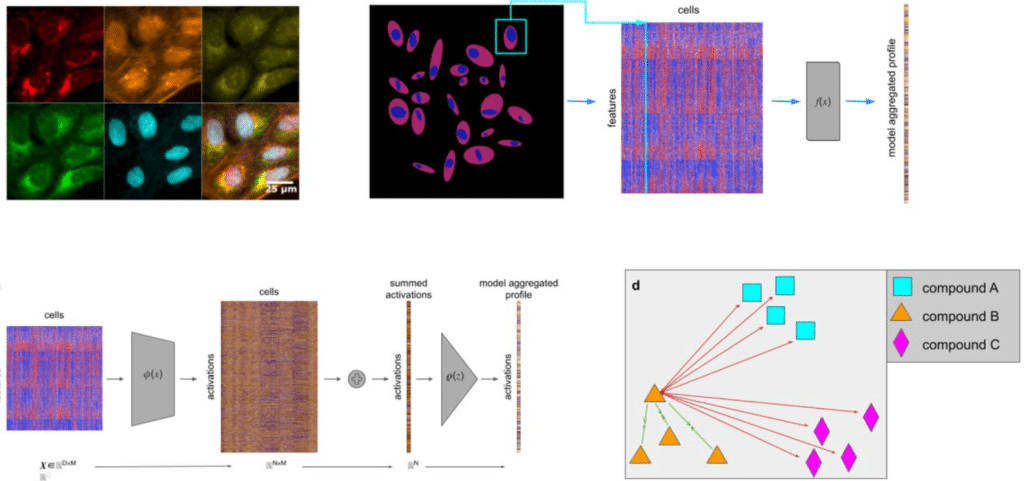
Capturing cell heterogeneity in representations of cell populations for image-based profiling using contrastive learning
Robert van Dijk, John Arevalo, Mehrtash Babadi, Anne E. Carpenter, Shantanu Singh
Phase-Restoring Subpixel Image Registration: Enhancing Motion Detection Performance in Fourier-domain Optical Coherence Tomography
Huakun Li, Bingyao Tan, Vimal Prabhu Pandiyan, Veluchamy Amutha Barathi, Ramkumar Sabesan, Leopold Schmetterer, Tong Ling
A Benchmark for Virus Infection Reporter Virtual Staining in Fluorescence and Brightfield Microscopy
Maria Wyrzykowska, Gabriel della Maggiora, Nikita Deshpande, Ashkan Mokarian, Artur Yakimovich
TUSCAN: Tumor segmentation and classification analysis in spatial transcriptomics
Chenxuan Zang, Charles C. Guo, Peng Wei, Ziyi Li
Visual analysis of spatial transcriptomics data with RedeViz
Dehe Wang, Xianwen Ren
Advances in Multiple Instance Learning for Whole Slide Image Analysis: Techniques, Challenges, and Future Directions
Jun Wang, Yu Mao, Nan Guan, Chun Jason Xue
HistoGym: A Reinforcement Learning Environment for Histopathological Image Analysis
Zhi-Bo Liu, Xiaobo Pang, Jizhao Wang, Shuai Liu, Chen Li


 (No Ratings Yet)
(No Ratings Yet)