Featured image with Jiwon Lee
Posted by FocalPlane, on 6 December 2024
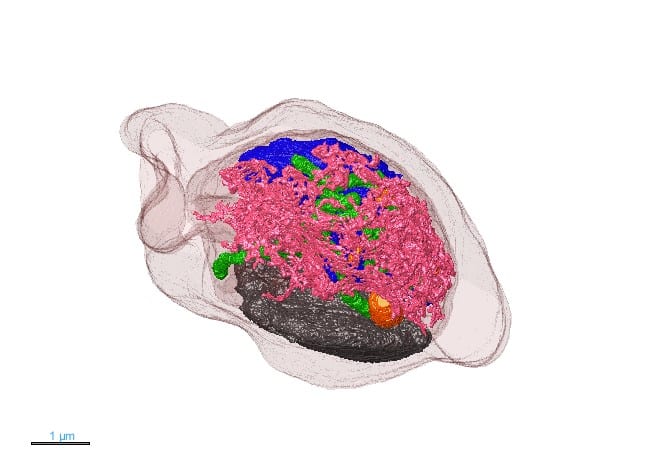
Our featured image, acquired by Jiwon Lee, is a 3D rendering model of Plasmodium falciparum showing the complex organisation of cellular organelles within the malaria parasite. Using a focused ion beam scanning electron microscope (FIB-SEM), a series of high resolution 2D images were acquired. The image stack was aligned and each organelle was segmented in the program MIB (Microscopy Image Browser) before the rendering process with Dragonfly software. This method provides nanometer scale resolution with a 3D perspective, enabling to study how organelles develop and how they interact with each other. Given the parasite’s small size, such intricate structural information is typically challenging to obtain, yet FIB-SEM’s 3D capabilities allow us to visualise and gain new insight into P. falciparum’s complex structure.
Find out more about Jiwon’s research below:
Research career so far: Throughout my career, I have worked extensively with a variety of microscopy techniques to support a wide range of research projects. This exposure has sparked a growing interest in cell biology, which I am currently exploring in my PhD research by studying lipid storage and trafficking mechanisms in malaria parasites. I am keen to continue investigating Plasmodium’s cellular architecture to deepen our understanding of its unique survival strategies.
Current research: Plasmodium falciparum is the deadliest species of malaria parasites infecting humans. It requires high lipid intake during the asexual blood stage to support growth. Our study, published in Journal of Cell Science, focuses on how the parasite stores and distributes lipids when needed. Using advanced microscopy techniques like FIB-SEM, we study the size, composition, and behaviour of lipid droplets (LDs) in the intraerythrocytic life cycle stages of the malaria parasite. Our findings revealed that LDs are crucial for lipid storage and homeostasis. Currently, we are exploring lipid metabolism and storage during other life cycle stages of Plasmodium falciparum.
Favourite imaging technique/microscope: Transmission electron microscopy (TEM) has always held a special place for me, despite the lengthy sample preparation, as it provides an incredible level of detail, making the process so rewarding. Recently, however, my work with FIB-SEM and electron tomography has shifted my focus toward volume EM techniques. These methods add a new dimension by combining high-resolution imaging with 3D insights, making volume EM my current favourite imaging technique for capturing complex cellular structures.
What are you most excited about in microscopy? The rapid advancements in visualisation tools, especially with AI-powered models, have made microscopy analysis significantly more efficient. At the start of my PhD, I manually segmented my data which took months. Now, AI allows me to create more accurate and visually compelling 3D models in much shorter time. I enjoy exploring various visualisation platforms to discover which best presents detailed information in an effective way to communicate complex biological concepts. The future of microscopy lies in these advanced visualisation tools in my view, offering more powerful ways to unravel and share the intricacies of biology.


 (No Ratings Yet)
(No Ratings Yet)