Probes 101 – How to choose a fluorescent probe for your imaging experiment
Posted by Lina El Hajji, on 14 December 2024
Fluorescence microscopy is a powerful tool for the visualization of the intricate biological processes occurring within biological organisms. Just like you can make important pieces of information pop up in their notes using colorful highlighters, biologists have at hand a large toolkit of “highlighters” to make their favorite targets stand out in the complex cellular environment. Attaching a fluorescent probe to a biomolecule of interest allows its direct visualization using optical microscopes, and the fluorescence signal gives precious information about its localization and levels as well as its interaction with potential partners in the cells. New challenges in the field of bioimaging arose from the need to answer evermore complex biological questions, ranging from the visualization of the finest architectural details of a given biological structure to the simultaneous detection of the interplay between multiple targets at the same time. So, how to choose the best fluorescent probe for a given imaging experiment?
Considerations on the photophysical properties of the probe
When selecting a suitable fluorescent probe for a given bioimaging context, there are a few considerations on its photophysical properties to bear in mind:
- the color of the reporter, meaning the wavelengths of absorption and emission are key determinants. These properties are important when wanting to visualize several targets at the same time in order to avoid having overlaps in their signals, and also depending on the depth at which the target is in the biological sample, as imaging in deep tissue often requires having near-infrared excitation and emission.
- the fluorescent lifetime of the reporter, meaning the average duration of the excited state of the fluorophore. This is a property which is often overlooked in fluorescent probes as we often tend to classify the probes in terms of color but the fluorescence lifetime is an additional signature of the reporter, which can also be leveraged in multiplexing experiments. An interesting feature of the fluorescence lifetime is that it is independent of the concentration of the fluorescence species. Thanks to improvements in hardware in fluorescence lifetime imaging microscopy (FLIM), multiplexing possibilities can be enhanced through combining detection based on the spectral properties to fluorescence lifetime signal.
- the brightness of the reporter ensures to have sufficient signal for imaging. When imaging scarce biological targets, having a bright probe ensures to have correct detection.
- the photostability of the reporter is an important property to keep in mind when needing long acquisition times or high imaging rate, or imaging with high laser power.
An ideal probe should thus be as bright as possible to maximize detection, cause minimal toxicity and damage to the sample, should have red-shifted spectral properties to allow deep-tissue imaging and display high photostability and high brightness to allow long timelapse imaging experiments and be suitable for super-resolution imaging modalities. Such a probe does not exist (yet)! For instance, higher photostability or red-shifted dyes usually come at the expense of high brightness. Better spatial resolution is obtained with blue-shifted wavelengths, however excitation in this region of the spectrum can cause higher phototoxicity. There is usually an interplay between all these properties, making the choice of the probe a compromise to make depending on the requirements of the experiment.
The nature of the probe matters
While the aforementioned properties are characteristics of all fluorescent reporters, there are several families of fluorescent systems that can be used in imaging experiments. Let’s first start with a quick and non-exhaustive overview on some of the big families of fluorescent probes: synthetic small molecule dyes and fluorescent proteins. On one hand, small molecule dyes are widely renowned for their excellent photophysical properties. Altering their structures greatly modifies their colors and properties, making them extremely versatile tools. Fluorescent dyes for imaging specific organelles of the cells have been developed, in additions to ones that do not get inside the cells for plasma membrane imaging. “Always on” dyes or dyes that turn on in specific environments have also been extensively engineered by chemists to improve their properties1. Dyes that blink have been developed for specific super-resolution imaging modalities2, bringing us snapshots of biological samples with an unprecedent level of details. While they are small in size, they often are linked with unspecific targeting, although molecular engineering efforts aim at improving this drawback.
On the other hand, fluorescent proteins have completely revolutionized the field of bioimaging. These are quite fascinating entities: some of their amino acids assemble into a chromophore that is fluorescent upon folding of the protein, well protected and shielded within its cavity3. The main asset of fluorescent proteins remains the high specificity of their targeting: the fluorescent protein gene is directly fused to the protein of interest gene, resulting in a chimeric protein after expression by the cellular machinery. Since the initial discovery and application of the green fluorescent protein in imaging experiments, extensive engineering efforts have enabled the development of a palette of fluorescent proteins, with different colors and properties, which have then been used to shine the light on a variety of biological processes, advancing our knowledge on how our cells function at a molecular level4. Although the use of these tools is widespread among biologists, their size and tendency to oligomerize perturb to some extent the visualized targets, as well as lacking the versatility of small molecule dyes. The chromophore formation reaction is also long, and requires molecular oxygen, hampering their use in oxygen deprived environments.
More recently, a new class of fluorescent reporters has emerged, trying to bridge the gap between synthetic dyes and genetically-encoded fluorescent tools, or combining “the best of both worlds” as recently reviewed by Robert E. Campbell5: chemogenetic fluorescent reporters. These hybrid reporters comprise a protein module that binds covalently or non-covalently to a fluorescently-labeled synthetic ligand. Having explained that, the advantages of these reporters are quite clear: they combine the labeling specificity of fluorescent proteins as the protein module is directly fused to the protein of interest, and versatility of small molecule dyes as the fluorescent moiety on the ligand can be engineered at will. Among these reporters, the self-labeling tags HaloTag6 and SNAP-tag7 have been adopted by many labs. The fluorescent moieties attached to the substrates of these proteins have been extensively engineered to improve brightness and photostability8,9, and protein engineering efforts have led to improving the labeling kinetics of these reporters10. Sensors and integrators of biological processes have also been designed using these strategies for labeling11. While HaloTag and SNAP-tag covalently bind their fluorescently-labeled substrates, other proteins scaffolds have been investigated since the groundbreaking work of Szent-Gyorgyi and Waggoner on developing fluorescence-activating proteins12. These proteins non-covalently interact with a small non-fluorescent ligand called a fluorogen, and this interaction leads to fluorescence activation of the fluorogen.
Our lab has contributed to this family of reporters through the development of the fluorescence-activating and absorption-shifting tag (FAST)13. FAST is a 14 kDa protein evolved through directed evolution from a bacterial photoreceptor protein called the photoactive yellow protein PYP to bind and activate the fluorescence of hydroxybenzylidene rhodanine (HBR) derivatives, which are push-pull molecules quite close in structure to the chromophore of the GFP13. Speaking of GFP, FAST is twice smaller than GFP, making it a potential less perturbative reporter. One great thing about these hybrid approaches is the possibility to tailor them either through engineering of the protein or molecular engineering of the fluorogen, or even both – which is often required. In the case of FAST, these engineering efforts have extended this family of reporters with new additions that are specifically tailored for given biological contexts14: a promiscuous tag that binds to fluorogens of different spectral properties opening a rainbow of imaging possibilities15, orthogonal reporters for multicolor imaging16, a near-infrared reporter for multiplexing experiments17, as well as fluorescence-lifetime modulating tags for in vivo multiplexing18.
Although these reporters hold great promise in combining advantages of genetically-encoded reporters and small molecule dyes, they are also linked to some of their limitations. Thus, efforts of these reporters should address the delivery of the fluorescent ligands or fluorogens in complex situations such as in small animals as well as reduction of their genetic footprint.
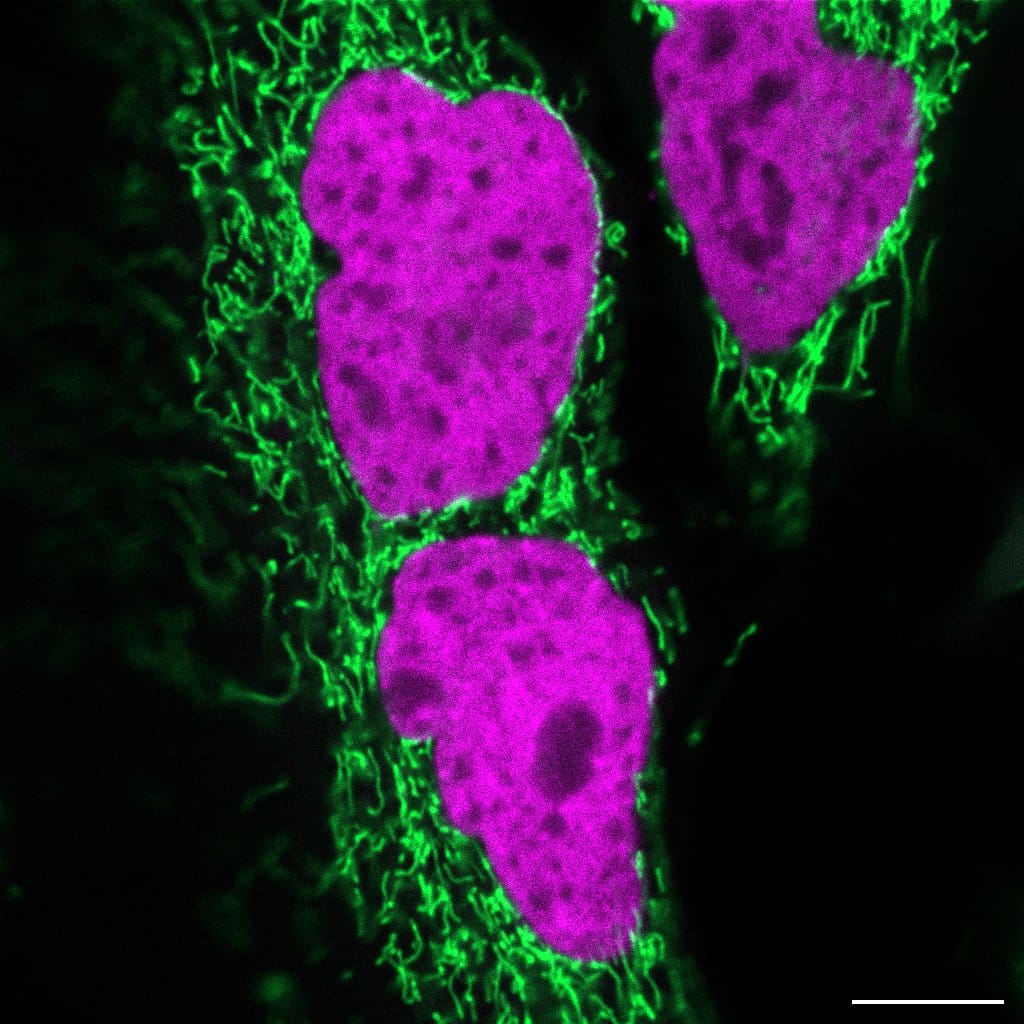
Figure 1: HeLa cells expressing mito-pFAST and H2B-nirFAST. Scale bar, 10 µm.
Do we still need more probes?
With all of these tools in hand, whether we still need to make new fluorescent probes is a legitimate question. When exploring older reviews on fluorescent reporters and the proposed directions on improvement19, many challenges have already been addressed: identifying and making new red shifted fluorescent proteins with higher brightness has been achieved through for instance the development of mCherry-XL20 and mScarlett321, improving the photostability of rhodamine dyes has been enabled by introduction of azetidine rings to the rhodamine structure9, and gaining photostability without expenses on brightness has been made possible by the development of StayGold protein and its latest variants22,23. These examples are all relatively recent which means that the field of fluorescent probes is still a promising research field. Since the initial developments of fluorescent probes, both imaging modalities and biological models used in experiments have greatly evolved, requiring the need for new and suitable probes with adequate properties to match the requirements of these evolutions.
In the last years, protein prediction tools as well as in silico docking and modelling softwares have emerged as an interesting alternative to designing improved fluorescent probes, as it can help bypass the longer engineering efforts required to modulate the structure of existing reporters by giving structural insights on specific hotspots to target. Although there are still very few examples of de novo designed fluorescent reporters24–26, it will be exciting to see what the future holds for this growing research area.
To finish, Rodriguez’s review on fluorescent proteins raised many questions regarding what still needs to be done in this field4. One drew my attention :
“Can we develop fluorescent protein variants to both observe and manipulate cellular functions simultaneously in the cell using fully optical approaches?”
Our lab has contributed to answering this question through the development of CATCHFIRE, which stands for chemically assisted tethering of chimera by fluorogenic-induced recognition27. The approach is interesting: with one tool, you can both induce a cellular process and visualize it with a fluorescent readout in real time. This tool has been applied to control protein proximity or induce organelle repositioning within cells. The development and improvement of such two-in-one strategies will deliver new tools for biologists to help advance our knowledge on cellular processes.
Harnessing knowledge on both chemistry, biology and photophysics and keeping up with the need to answer evermore complex biological questions has enabled the construction of a large toolkit of available fluorescent reporters, with improved properties and applicability, and the field is a nice illustration of how important interdisciplinary research is. Whether it is through using dyes or fluorescent proteins or hybrid approaches, we are surely not done shining light on biological organisms to make beautiful and colorful snapshots illustrating nature’s complexity.
Lina El Hajji Postdoctoral researcher at Sorbonne University
References
1. Grimm, J. B. & Lavis, L. D. Caveat fluorophore: an insiders’ guide to small-molecule fluorescent labels. Nature Methods 19, 149–158 (2022).
2. Aparin, I. O., Yan, R., Pelletier, R., Choi, A. A., Danylchuk, D. I., Xu, K. & Klymchenko, A. S. Fluorogenic Dimers as Bright Switchable Probes for Enhanced Super-Resolution Imaging of Cell Membranes. Journal of the American Chemical Society 144, 18043–18053 (2022).
3. Tsien, R. Y. The green fluorescent protein. Annual Review of Biochemistry 67, 509–544 (1998).
4. Rodriguez, E. A., Campbell, R. E., Lin, J. Y., Lin, M. Z., Miyawaki, A., Palmer, A. E., Shu, X., Zhang, J. & Tsien, R. Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends in Biochemical Sciences 42, 111–129 (2017).
5. Tsao, K. K., Imai, S., Chang, M., Hario, S., Terai, T. & Campbell, R. E. The best of both worlds: Chemigenetic fluorescent sensors for biological imaging. Cell Chemical Biology 31, 1652–1664 (2024).
6. Los, G. V., Encell, L. P., McDougall, M. G., Hartzell, D. D., Karassina, N., Zimprich, C., Wood, M. G., Learish, R., Ohana, R. F., Urh, M., Simpson, D., Mendez, J., Zimmerman, K., Otto, P., Vidugiris, G., Zhu, J., Darzins, A., Klaubert, D. H., Bulleit, R. F. & Wood, K. V. HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chemical Biology 3, 373–382 (2008).
7. Juillerat, A., Gronemeyer, T., Keppler, A., Gendreizig, S., Pick, H., Vogel, H. & Johnsson, K. Directed Evolution of O6-Alkylguanine-DNA Alkyltransferase for Efficient Labeling of Fusion Proteins with Small Molecules In Vivo. Chemistry & Biology 10, 313–317 (2003).
8. Grimm, J. B., Muthusamy, A. K., Liang, Y., Brown, T. A., Lemon, W. C., Patel, R., Lu, R., Macklin, J. J., Keller, P. J., Ji, N. & Lavis, L. D. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nature Methods 14, 987–994 (2017).
9. Grimm, J. B., English, B. P., Chen, J., Slaughter, J. P., Zhang, Z., Revyakin, A., Patel, R., Macklin, J. J., Normanno, D., Singer, R. H., Lionnet, T. & Lavis, L. D. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nature Methods 12, 244–250 (2015).
10. Kühn, S., Nasufovic, V., Wilhelm, J., Kompa, J., de Lange, E. M. F., Lin, Y.-H., Egoldt, C., Fischer, J., Lennoi, A., Tarnawski, M., Reinstein, J., Vlijm, R., Hiblot, J. & Johnsson, K. SNAP-tag2: faster and brighter protein labeling. bioRxiv 2024.08.28.610127 (2024). doi:10.1101/2024.08.28.610127
11. Huppertz, M.-C., Wilhelm, J., Grenier, V., Schneider, M. W., Falt, T., Porzberg, N., Hausmann, D., Hoffmann, D. C., Hai, L., Tarnawski, M., Pino, G., Slanchev, K., Kolb, I., Acuna, C., Fenk, L. M., Baier, H., Hiblot, J. & Johnsson, K. Recording physiological history of cells with chemical labeling. at <https://www.science.org>
12. Szent-Gyorgyi, C., Schmidt, B. A., Creeger, Y., Fisher, G. W., Zakel, K. L., Adler, S., Fitzpatrick, J. A. J., Woolford, C. A., Yan, Q., Vasilev, K. V., Berget, P. B., Bruchez, M. P., Jarvik, J. W. & Waggoner, A. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nature Biotechnology 26, 235–240 (2008).
13. Plamont, M. A., Billon-Denis, E., Maurin, S., Gauron, C., Pimenta, F. M., Specht, C. G., Shi, J., Quérard, J., Pan, B., Rossignol, J., Morellet, N., Volovitch, M., Lescop, E., Chen, Y., Triller, A., Vriz, S., Saux, T. L., Jullien, L. & Gautier, A. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proceedings of the National Academy of Sciences of the United States of America 113, 497–502 (2016).
14. Gautier, A. Fluorescence-Activating and Absorption-Shifting Tags for Advanced Imaging and Biosensing. Accounts of Chemical Research 55, 3125–3135 (2022).
15. Benaissa, H., Ounoughi, K., Aujard, I., Fischer, E., Goïame, R., Nguyen, J., Tebo, A. G., Li, C., Saux, T. L., Bertolin, G., Tramier, M., Danglot, L., Pietrancosta, N., Morin, X., Jullien, L. & Gautier, A. Engineering of a fluorescent chemogenetic reporter with tunable color for advanced live-cell imaging. Nature Communications 12, 6989 (2021).
16. Tebo, A. G., Moeyaert, B., Thauvin, M., Carlon-Andres, I., Böken, D., Volovitch, M., Padilla-Parra, S., Dedecker, P., Vriz, S. & Gautier, A. Orthogonal fluorescent chemogenetic reporters for multicolor imaging. Nature Chemical Biology 17, 30–38 (2021).
17. El Hajji, L., Bunel, B., Joliot, O., Li, C., Tebo, A. G., Rampon, C., Volovitch, M., Fischer, E., Pietrancosta, N., Perez, F., Morin, X., Vriz, S. & Gautier, A. A tunable and versatile chemogenetic near infrared fluorescent reporter. biorXiv (2024). doi:10.1101/2024.04.05.588310
18. El Hajji, L., Lam, F., Avtodeeva, M., Benaissa, H., Rampon, C., Volovitch, M., Vriz, S. & Gautier, A. Multiplexed In Vivo Imaging with Fluorescence Lifetime-Modulating Tags. Advanced Science 11, 2404354 (2024).
19. Zhang, J., Campbell, R. E., Ting, A. Y. & Tsien, R. Y. Creating new fluorescent probes for cell biology. Nature Reviews Molecular Cell Biology 3, 906–918 (2002).
20. Mukherjee, S., Manna, P., Hung, S. T., Vietmeyer, F., Friis, P., Palmer, A. E. & Jimenez, R. Directed Evolution of a Bright Variant of mCherry: Suppression of Nonradiative Decay by Fluorescence Lifetime Selections. Journal of Physical Chemistry B 126, 4659–4668 (2022).
21. Gadella, T. W. J., Weeren, L. van, Stouthamer, J., Hink, M. A., Wolters, A. H. G., Giepmans, B. N. G., Aumonier, S., Dupuy, J. & Royant, A. mScarlet3: a brilliant and fast-maturing red fluorescent protein. Nature Methods 20, 541–545 (2023).
22. Hirano, M., Ando, R., Shimozono, S., Sugiyama, M., Takeda, N., Kurokawa, H., Deguchi, R., Endo, K., Haga, K., Takai-Todaka, R., Inaura, S., Matsumura, Y., Hama, H., Okada, Y., Fujiwara, T., Morimoto, T., Katayama, K. & Miyawaki, A. A highly photostable and bright green fluorescent protein. Nature biotechnology (2022). doi:10.1038/s41587-022-01278-2
23. Goedhart, J. & Gadella, T. W. J. Breaking up the StayGold dimer yields three photostable monomers. Nature Methods 21, 558–559 (2024).
24. Dou, J., Vorobieva, A. A., Sheffler, W., Doyle, L. A., Park, H., Bick, M. J., Mao, B., Foight, G. W., Lee, M. Y., Gagnon, L. A., Carter, L., Sankaran, B., Ovchinnikov, S., Marcos, E., Huang, P.-S., Vaughan, J. C., Stoddard, B. L. & Baker, D. De novo design of a fluorescence-activating β-barrel. Nature 561, 485–491 (2018).
25. Klima, J. C., Doyle, L. A., Lee, J. D., Rappleye, M., Gagnon, L. A., Lee, M. Y., Barros, E. P., Vorobieva, A. A., Dou, J., Bremner, S., Quon, J. S., Chow, C. M., Carter, L., Mack, D. L., Amaro, R. E., Vaughan, J. C., Berndt, A., Stoddard, B. L. & Baker, D. Incorporation of sensing modalities into de novo designed fluorescence-activating proteins. Nature Communications 12, 1–19 (2021).
26. Baker, D., Xu, C., Liu, Y., Arús, B., Mishra, K., Luciano, M., Bandi, V., Bick, M., Xu, M., Zhang, K., Helmholtz, J. L., München, Z., Bae, J., Kang, A. & Gerben, S. De novo Design of Near Infrared Fluorescent Proteins. doi:10.21203/rs.3.rs-4652998/v1
27. Bottone, S., Joliot, O., Cakil, Z. V., El Hajji, L., Rakotoarison, L. M., Boncompain, G., Perez, F. & Gautier, A. A fluorogenic chemically induced dimerization technology for controlling, imaging and sensing protein proximity. Nature Methods 20, 1553–1562 (2023).


 (No Ratings Yet)
(No Ratings Yet)
