Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 18 April 2025
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
F-BIAS: Towards a distributed national core facility for Bioimage Analysis
Mélodie Ambroset, Marie Anselmet, Clément Benedetti, Arthur Meslin, Aurélien Maillot, Christian Rouvière, Guillaume Maucort, Marine Breuilly, Marvin Albert, Gaëlle Letort, Antonio Trullo, Volker Bäcker, Jérôme Mutterer, Fabrice Cordelières, Thierry Pécot, Minh-Son Phan, Stéphane Rigaud, Magali Feyeux, Perrine Paul-Gilloteaux, Anne-Sophie Macé, Jean-Yves Tinevez
2024 OME-NGFF workflows hackathon
Joel Lüthi, Marvin Albert, Liviu Anita, Kola Babalola, Davis Bennett, John A. Bogovic, Lorenzo Cerrone, Rémy Dornier, Jan Eglinger, Vera Galinova, Reto Gerber, Oane Gros, Stefan Hahmann, Max Hess, Ruth Hornbachner, Dmytro Horyslavets, Rachael Huxford, Daniel Krentzel, Tong LI, Luca Marconato, Matthew McCormick, Franziska Moos, Filip Mroz, Bugra Özdemir, Benjamin Pavie, Eric Perlman, Maximilian Schulz, Leonardo Schwarz, Hannes M. Spitz, David Stansby, Fabio Steffen, Szymon Stoma, Flurin Sturzenegger, Wouter-Michiel Vierdag, Jonas Windhager, Kevin Yamauchi, Igor Zubarev, Josh Moore, Norman Rzepka, Christian Tischer, Vladimir Ulman, and Virginie Uhlmann
Using Beads as a Focus Fiduciary to Aid Software-based Autofocus Accuracy in Microscopy
I. Gibson, E.J. Osterlund, R. Truant
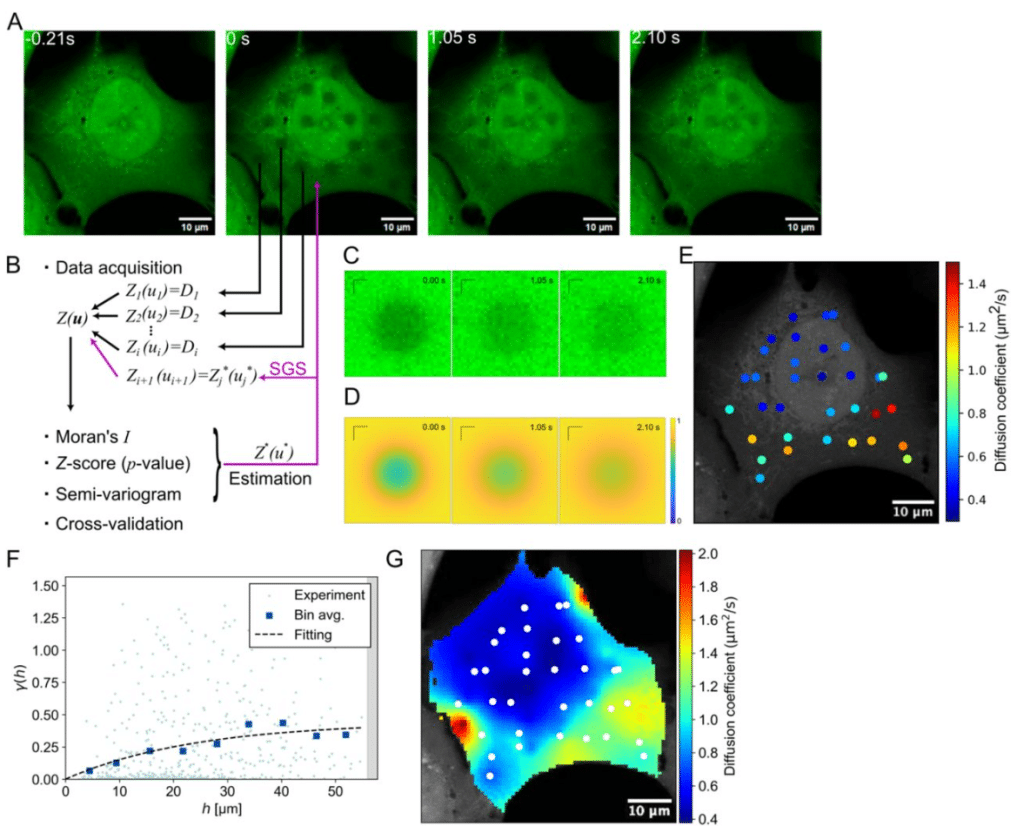
Mapping molecular diffusion across the whole cell with spatial statistics-based FRAP
Yohei Okabe, Takumi Saito, Outa Nakashima, Daiki Matsunaga, Shinji Deguchi
Counting particles could give wrong probabilities in Cryo-Electron Microscopy
Luke Evans, Lars Dingeldein, Roberto Covino, Marc Aurèle Gilles, Erik Thiede, Pilar Cossio
FRAP analysis Measuring biophysical kinetic parameters using image analysis
Sharva V. Hiremath, Etika Goyal, Gregory T. Reeves, Cranos M. Williams
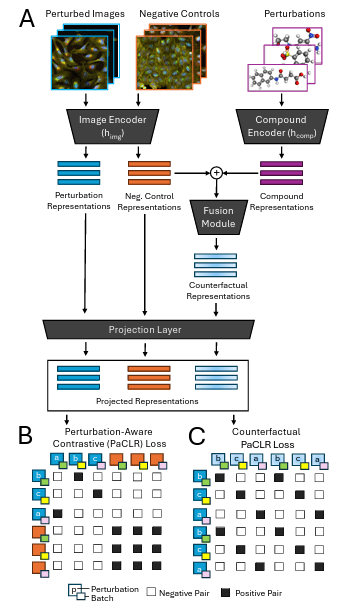
Causal integration of chemical structures improves representations of microscopy images for morphological profiling
Yemin Yu, Neil Tenenholtz, Lester Mackey, Ying Wei, David Alvarez-Melis, Ava P. Amini, Alex X. Lu
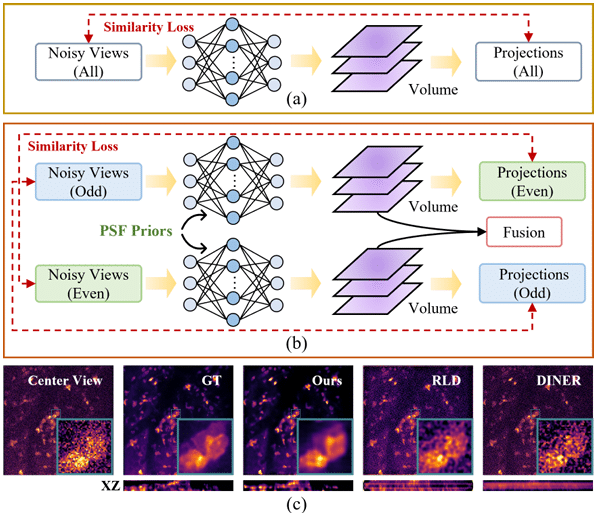
V2V3D: View-to-View Denoised 3D Reconstruction for Light-Field Microscopy
Jiayin Zhao, Zhenqi Fu, Tao Yu, Hui Qiao
Inferring scattering-type Scanning Near-Field Optical Microscopy Data from Atomic Force Microscopy Images
Stefan G. Stanciu, Stefan R. Anton, Denis E. Tranca, George A. Stanciu, Bogdan Ionescu, Zeev Zalevsky, Binyamin Kusnetz, Jeremy Belhassen, Avi Karsenty, Gabriella Cincotti
indiSplit: Bringing Severity Cognizance to Image Decomposition in Fluorescence Microscopy
Ashesh Ashesh, Florian Jug
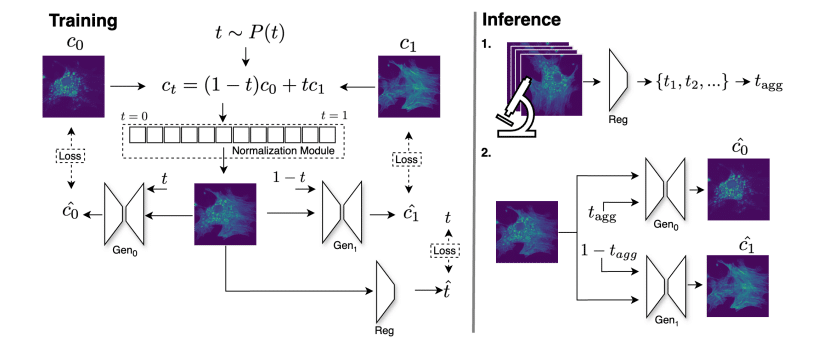
RxRx3-core: Benchmarking drug-target interactions in High-Content Microscopy
Oren Kraus, Federico Comitani, John Urbanik, Kian Kenyon-Dean, Lakshmanan Arumugam, Saber Saberian, Cas Wognum, Safiye Celik, Imran S. Haque
Out-of-distribution evaluations of channel agnostic masked autoencoders in fluorescence microscopy
Christian John Hurry, Jinjie Zhang, Olubukola Ishola, Emma Slade, Cuong Q. Nguyen
Adapting Video Diffusion Models for Time-Lapse Microscopy
Alexander Holmberg, Nils Mechtel, Wei Ouyang
3-D Image-to-Image Fusion in Lightsheet Microscopy by Two-Step Adversarial Network: Contribution to the FuseMyCells Challenge
Marek Wodzinski, Henning Müller
Zero-Shot Denoising for Fluorescence Lifetime Imaging Microscopy with Intensity-Guided Learning
Hao Chen, Julian Najera, Dagmawit Geresu, Meenal Datta, Cody Smith, Scott Howard
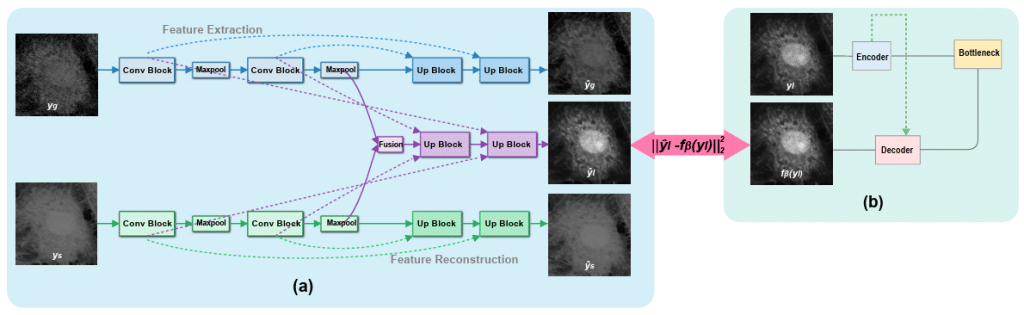
MicroVQA: A Multimodal Reasoning Benchmark for Microscopy-Based Scientific Research
James Burgess, Jeffrey J Nirschl, Laura Bravo-Sánchez, Alejandro Lozano, Sanket Rajan Gupte, Jesus G. Galaz-Montoya, Yuhui Zhang, Yuchang Su, Disha Bhowmik, Zachary Coman, Sarina M. Hasan, Alexandra Johannesson, William D. Leineweber, Malvika G Nair, Ridhi Yarlagadda, Connor Zuraski, Wah Chiu, Sarah Cohen, Jan N. Hansen, Manuel D Leonetti, Chad Liu, Emma Lundberg, Serena Yeung-Levy
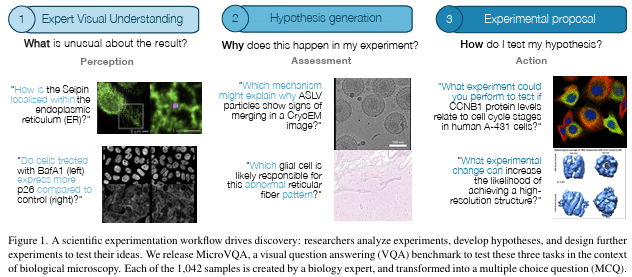
Enhancing Deep Learning Based Structured Illumination Microscopy Reconstruction with Light Field Awareness
Long-Kun Shan, Ze-Hao Wang, Tong-Tian Weng, Xiang-Dong Chen, Fang-Wen Sun
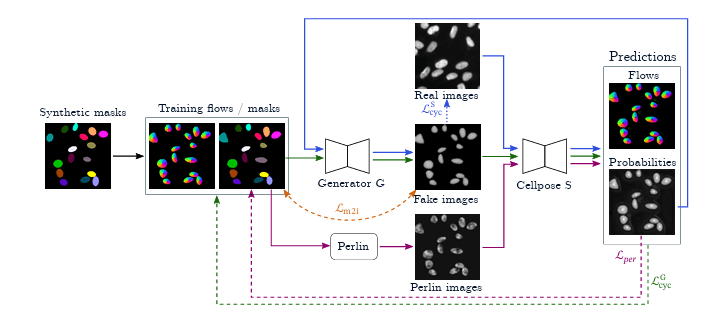
CyclePose — Leveraging Cycle-Consistency for Annotation-Free Nuclei Segmentation in Fluorescence Microscopy
Jonas Utz, Stefan Vocht, Anne Tjorven Buessen, Dennis Possart, Fabian Wagner, Mareike Thies, Mingxuan Gu, Stefan Uderhardt, Katharina Breininger
A Bi-channel Aided Stitching of Atomic Force Microscopy Images
Huanhuan Zhao, Ruben Millan-Solsona, Marti Checa, Spenser R. Brown, Jennifer L. Morrell-Falvey, Liam Collins, Arpan Biswas
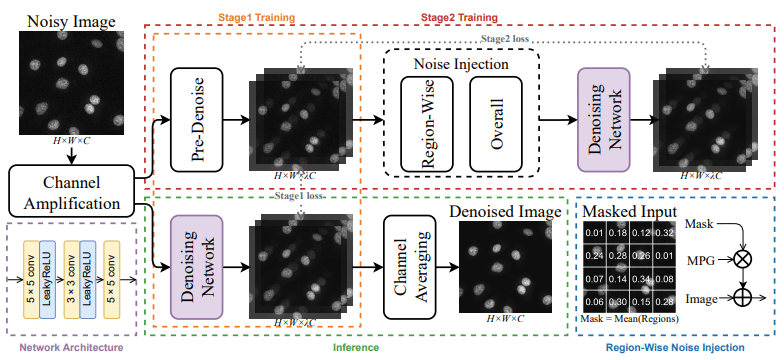
FM2S: Towards Spatially-Correlated Noise Modeling in Zero-Shot Fluorescence Microscopy Image Denoising
Jizhihui Liu, Qixun Teng, Qing Ma, Junjun Jiang
Enhancing Cell Instance Segmentation in Scanning Electron Microscopy Images via a Deep Contour Closing Operator
Florian Robert, Alexia Calovoulos, Laurent Facq, Fanny Decoeur, Etienne Gontier, Christophe F. Grosset, Baudouin Denis de Senneville
From Pixels to Histopathology: A Graph-Based Framework for Interpretable Whole Slide Image Analysis
Alexander Weers, Alexander H. Berger, Laurin Lux, Peter Schüffler, Daniel Rueckert, Johannes C. Paetzold
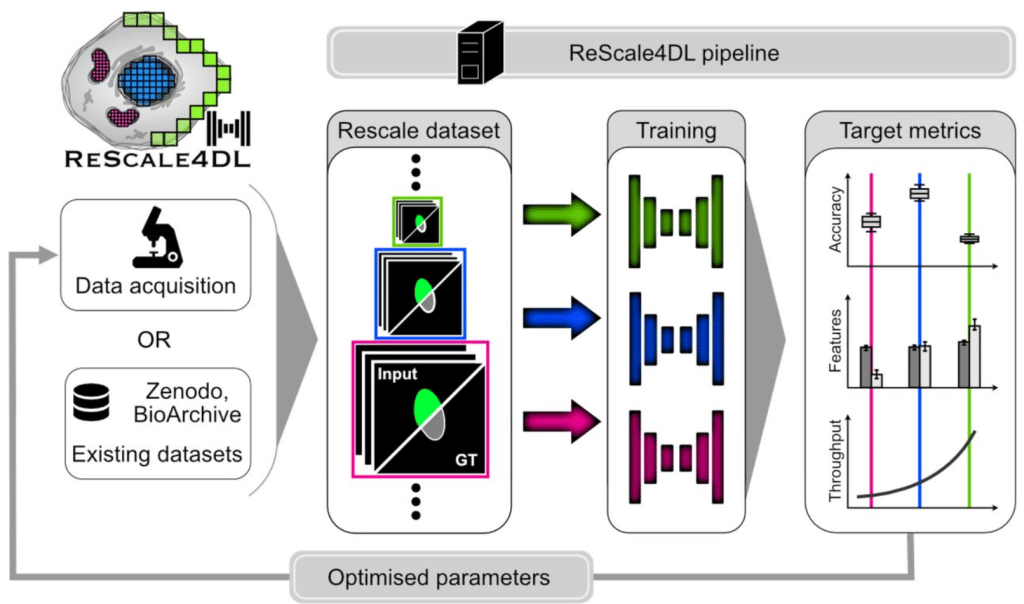
ReScale4DL: Balancing Pixel and Contextual Information for Enhanced Bioimage Segmentation
Mariana G. Ferreira, Bruno M. Saraiva, António D. Brito, Mariana G. Pinho, Ricardo Henriques, Estibaliz Gómez-de-Mariscal
DANEELpath: Open-source Digital analysis tools for histopathological research. Applications in NEuroblastoma modELs
Isaac Vieco-Martí, Amparo López-Carrasco, Samuel Navarro, Sofia Granados Aparici, Rosa Noguera
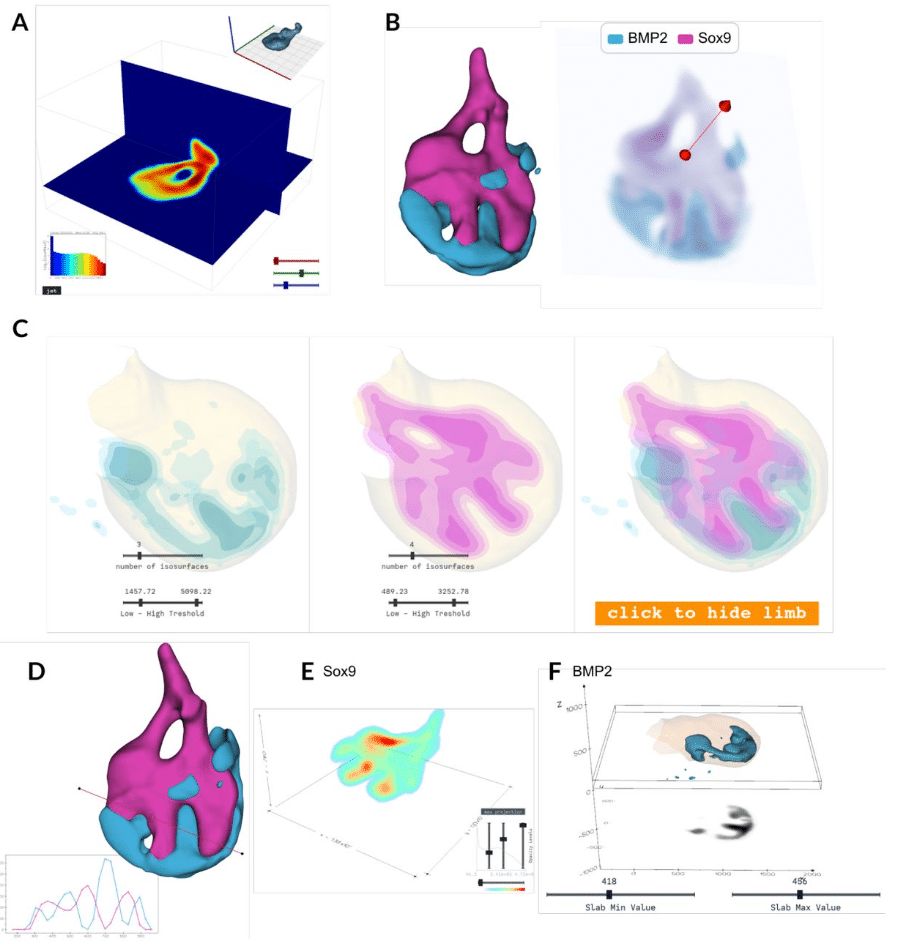
LimbLab: Pipeline for 3D Analysis and Visualisation of Limb Bud Gene Expression
Laura Aviñó-Esteban, Heura Cardona-Blaya, Marco Musy, Antoni Matyjaszkiewicz, James Sharpe, Giovanni Dalmasso
InSituPy – A framework for histology-guided, multi-sample analysis of single-cell spatial transcriptomics data
Johannes Wirth, Anna Chernysheva, Birthe Lemke, Isabel Giray, Aitana Egea Lavandera, Katja Steiger
VESNA: An Open-Source Tool for Automated 3D Vessel Segmentation and Network Analysis
Magdalena Schüttler, Leyla Doğan, Jana Kirchner, Süleyman Ergün, Philipp Wörsdörfer, Sabine C. Fischer
VNC-Dist: A machine learning-based pipeline for quantification of neuronal positioning in the ventral nerve cord of C. elegans
Saber Saharkhiz, Mearhyn Petite, Tony Roenspies, Theodore J. Perkins, Antonio Colavita
BRAPH 2: a flexible, open-source, reproducible, community-oriented, easy-to-use framework for network analyses in neurosciences
Yu-Wei Chang, Blanca Zufiria-Gerbolés, Emiliano Gómez-Ruiz, Anna Canal-García, Hang Zhao, Mite Mijalkov, Joana B. Pereira, Giovanni Volpe
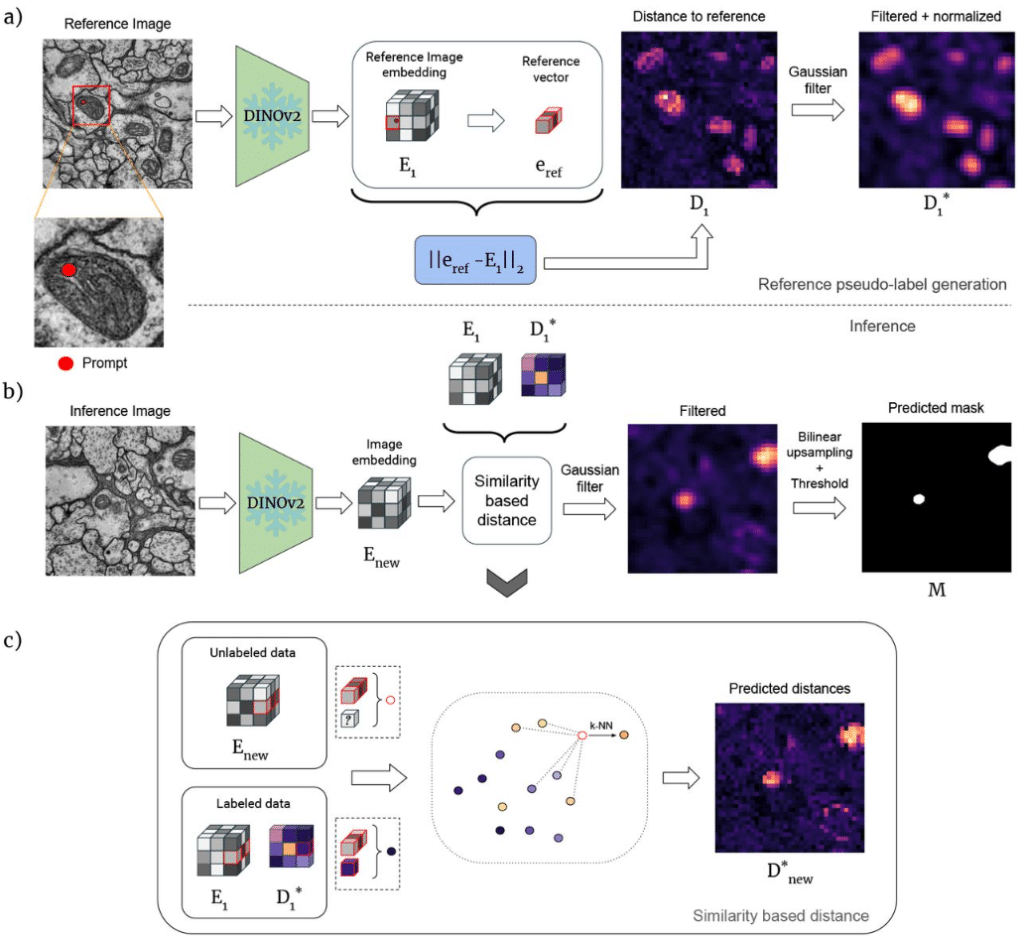
DINOSim: Zero-Shot Object Detection and Semantic Segmentation on Electron Microscopy Images
Aitor González-Marfil, Estibaliz Gómez-de-Mariscal, Ignacio Arganda-Carreras
Tracking Coordinated Cellular Dynamics in Time-Lapse Microscopy with ARCOS.px
Benjamin Grädel, Lea Brönnimann, Paolo Armando Gagliardi, Lucien Hinderling, Olivier Pertz, Maciej Dobrzyński
niiv: Interactive Self-supervised Neural Implicit Isotropic Volume Reconstruction
Jakob Troidl, Yiqing Liang, Johanna Beyer, Mojtaba Tavakoli, Johann Danzl, Markus Hadwiger, Hanspeter Pfister, James Tompkin
Label-free droplet image analysis with Cellprofiler
Dániel Kácsor, Merili Saar-Abroi, Triini Olman, Simona Bartkova, Ott Scheler
A guide for Single Particle Tracking: from sample preparation and image acquisition to the analysis of individual trajectories
Asaki Kobayashi, Junwoo Park, Judith Miné-Hattab, Fabiola García Fernández
StainStyleSampler: Clustering-based sampling of whole slide image appearances
Maya Maya Barbosa Silva, Sabine Leh, Hrafn Weishaupt
Helicon: Helical indexing and 3D reconstruction from one image
Daoyi Li, Xiaoqi Zhang, Wen Jiang
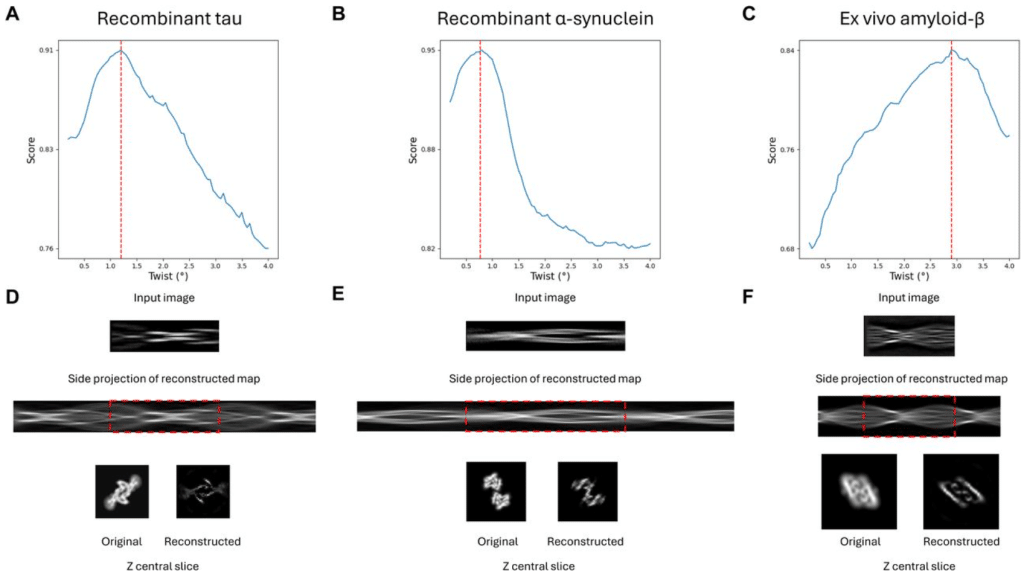
IntegrAlign: A comprehensive tool for multiimmunofluorescence panel integration through image alignment
Leo Hermet, Leo Laoubi, Martial Scavino, Anne-Claire Doffin, Alexia Gazeu, Berthet Justine, Berenice Pillat, Stephanie Tissot, Sylvie Rusakiewicz, Marie-Cecile Michallet, Nathalie Bendriss-Vermare, Jenny Valladeau-Guilemond, Jean Hausser, Christophe Caux, Margaux Hubert
Supervised Deep Learning for Efficient Cryo-EM Image Alignment in Drug Discovery with cryoPARES
Ruben Sanchez-Garcia, Alex Berndt, Amir Apelbaum, Judith Reeks, Pamela A Williams, Carl Poelking, Charlotte M Deane, Michael Saur
RNA2seg: a generalist model for cell segmentation in image-based spatial transcriptomics
Thomas Defard, Alice Blondel, Sebastien Bellow, Anthony Coleon, Guilherme Dias de Melo, Thomas Walter, Florian Mueller
SpatioCell: A Deep Learning Algorithm for High-resolution Single-cell Mapping through Deep Integration of Histology Image and Sequencing Data
Naiqiao Hou, Zhaorun Wu, Yue Yu, Yanqi Zhang, Jiayun Wu, Zhixing Zhong, Fangfang Xu, Zeyu Wang, Chaoyong Yang, Weihong Tan, Jia Song
A multi-modal whole-slide image processing pipeline for quantitative mapping of tissue architecture, histopathology, and tissue microenvironment
Maomao Chen, Hongqiang Ma, Xuejiao Sun, Marc Schwartz, Randall E. Brand, Jianquan Xu, Dimitrios S. Gotsis, Phuong Nguyen, Beverley A. Moore, Lori Snyder, Rhonda M. Brand, Yang Liu
MitoStructSeg: A Comprehensive Platform for Mitochondrial Structure Segmentation and Analysis
Xinsheng Wang, Buqing Cai, Zhuo Jia, Yuanbo Chen, Shuai Guo, Zheng Liu, Fuwei Li, Hao Dong, Fa Zhang, Xiaohua Wan, Bin Hu
OPUS-TOMO: Deep Learning Framework for Structural Heterogeneity Analysis in Cryo-electron Tomography
Zhenwei Luo, Xiangru Chen, Qinghua Wang, Jianpeng Ma
CABaNe, an automated, high content ImageJ macro for cell and neurite analysis
Nathan Thibieroz, João Lopes, Paul Machillot, Fabrice Cordelières, Catherine Picart, Elisa Migliorini
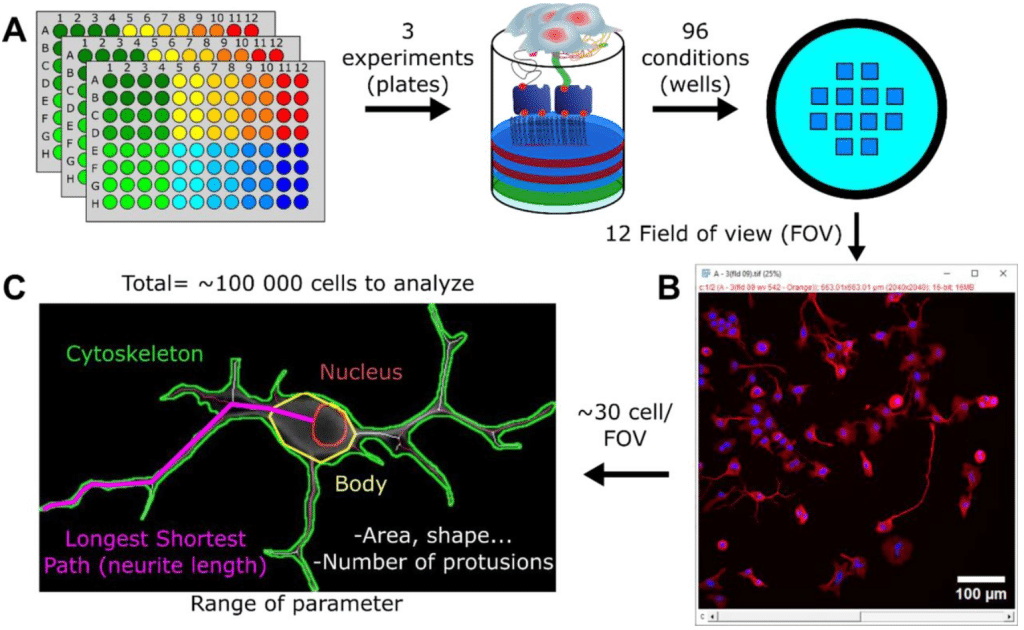
Suite3D: Volumetric cell detection for two-photon microscopy
Ali Haydaroğlu, Sam Dodgson, Michael Krumin, Andrew Landau, Liad J. Baruchin, Tinya Chang, Jingkun Guo, David Meyer, Charu Bai Reddy, Jian Zhong, Na Ji, Sylvia Schröder, Kenneth D Harris, Alipasha Vaziri, Matteo Carandini
Whole-cell particle-based digital twin simulations from 4D lattice light-sheet microscopy data
Eric Arkfeld, Zichen Wang, Hiroyuki Hakozaki, Johannes Schöneberg
Quantifying Uncertainty in Phasor-Based Time-Domain Fluorescence Lifetime Imaging Microscopy
Qinyi Chen, Jongchan Park, Shuqi Mu, Liang Gao


 (No Ratings Yet)
(No Ratings Yet)