Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 30 May 2025
Here is a curated selection of preprints published recently (up until 22 May). In this post, we focus specifically on bioimage analysis and data management.
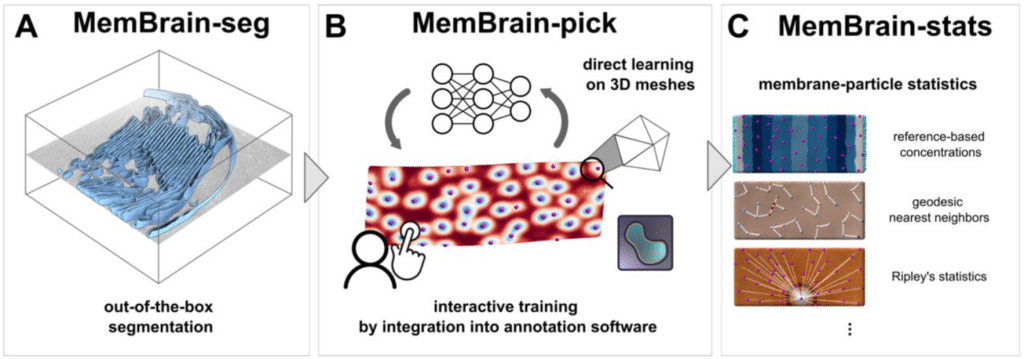
MemBrain v2: an end-to-end tool for the analysis of membranes in cryo-electron tomography
Lorenz Lamm, Simon Zufferey, Hanyi Zhang, Ricardo D. Righetto, Florent Waltz, Wojciech Wietrzynski, Kevin A. Yamauchi, Alister Burt, Ye Liu, Antonio Martinez-Sanchez, Sebastian Ziegler, Fabian Isensee, Julia A. Schnabel, Benjamin D. Engel, Tingying Peng
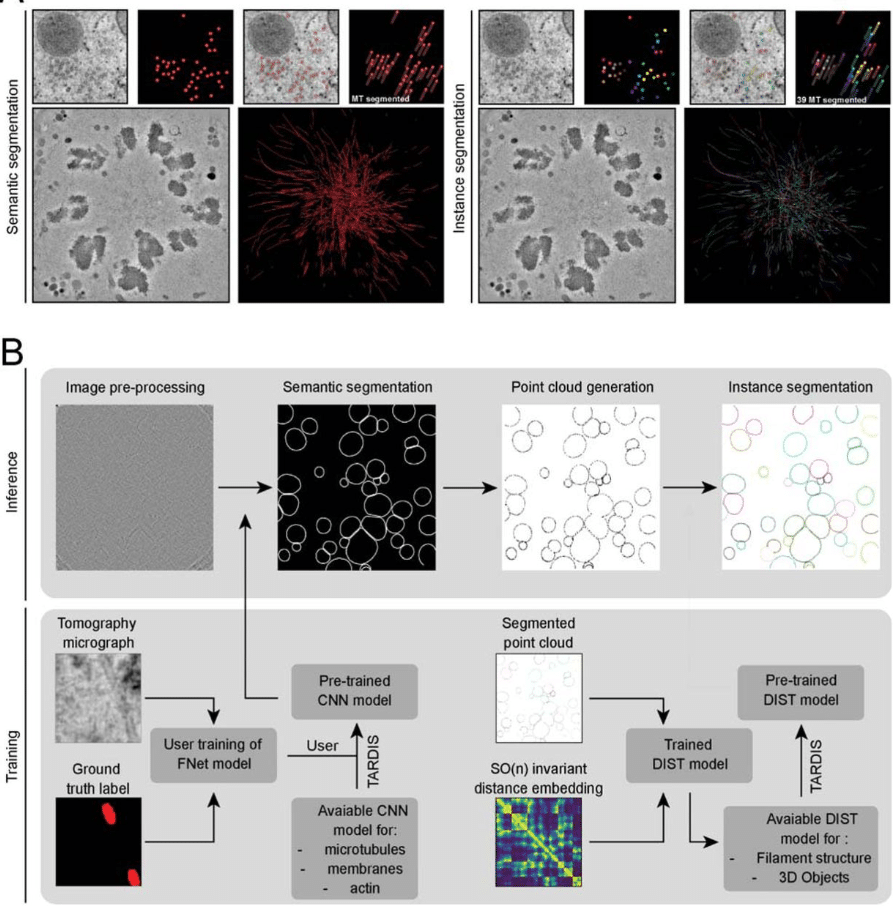
OmniEM: Unifying the EM Multiverse through a Large-scale Foundation Model
Liuyuan He, Ruohua Shi, Wenyao Wang, Lei Ma
AI4CellFate: Interpretable Early Cell Fate Prediction with Generative AI
Inês Cunha, Luca Panconi, Sebastian Bauer, Maxime Gestin, Emma Latron, Erik Sahai, Alix Le Marois, Juliette Griffié
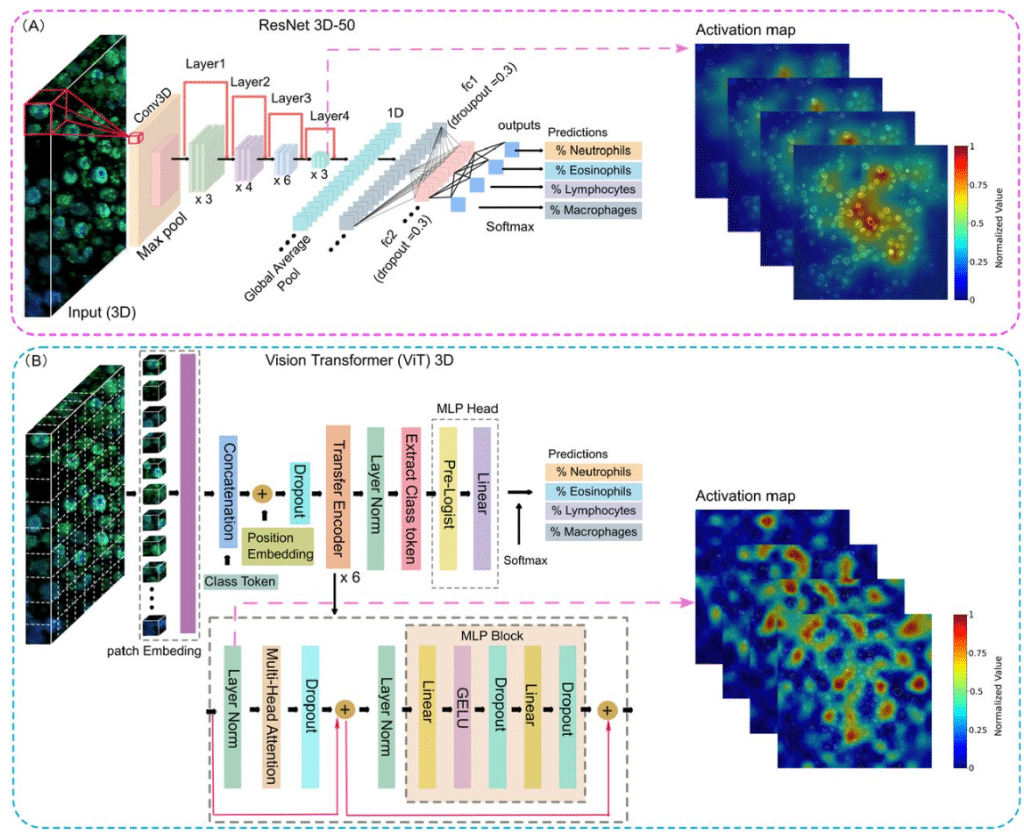
Deep Learning-Based 3D Leukocyte Differentiation Using Label-Free Higher Harmonic Generation Microscopy
Mengyao Zhou, Patrick José González, Tamara Dekker, Shiqi Zhang, Leonoor S. Boers, Hélène B. van den Heuvel, Annemiek Dijkhuis, Iris A. Simons, Jan Willem Duitman, Marie Louise Groot
A Comprehensive Workflow for Imaging Live Insulin Secretion Events and Granules in Intact Islets
Margret A. Fye, Rahul Sharma, Phyu Sin M. Myat, Pi’ilani Regan, Hudson McKinney, Guoqiang Gu, Irina Kaverina
Light-field deep learning enables high-throughput, scattering-mitigated calcium imaging
Carmel L. Howe, Kate L.Y. Zhao, Herman Verinaz-Jadan, Pingfan Song, Samuel J. Barnes, Pier Luigi Dragotti, Amanda J. Foust
Optical Phenotyping Using Label-Free Microscopy and Deep Learning
Shuyuan Guan, Thomas Knapp, Alba Alfonso-Garcia, Suzann Duan, Travis W. Sawyer
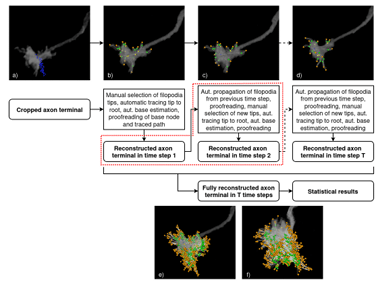
Semi-automatic Geometrical Reconstruction and Analysis of Filopodia Dynamics in 4D Two-Photon Microscopy Images
Blaz Brence, Josephine Brummer, Vincent J. Dercksen, Mehmet Nesel Ozel, Abhishek Kulkarni, Neele Wolterhoff, Steffen Prohaska, Peter Robin Hiesinger, Daniel Baum
A Multimodal Imaging Pipeline for the Discovery of Molecular Markers of Cellular Neighborhoods
Thai Pham, Melissa A. Farrow, Lukasz G. Migas, Martin Dufresne, Madeline E. Colley, Jamie L. Allen, Mark P. de Caestecker, Raf Van de Plas, Jeffrey M. Spraggins
Self-contrastive learning enables interference-resilient and generalizable fluorescence microscopy signal detection without interference modeling
Fengdi Zhang, Ruqi Huang, Meiqian Xin, Haoran Meng, Danheng Gao, Ying Fu, Juntao Gao, Xiangyang Ji
Universal Transformer-Based Tracker for Accurate Tracking of Particles and Cells in Microscopy
Yudong Zhang, Dan Liu, Ge Yang
A Deep Learning approach for time-consistent cell cycle phase prediction from microscopy data
Thomas Bonte, Oriane Pourcelot, Adham Safieddine, Floric Slimani, Florian Mueller, Dominique Weil, Edouard Bertrand, Thomas Walter
SoftPQ: Robust Instance Segmentation Evaluation via Soft Matching and Tunable Thresholds
Ranit Karmakar, Simon F. Nørrelykke
PyReconstruct: A fully opensource, collaborative successor to Reconstruct
Michael A. Chirillo, Julian N. Falco, Michael D. Musslewhite, Larry F. Lindsey, Kristen M. Harris
Grid partitioning image analysis for bacterial cell aggregates
Yuki Ohara, Shogo Yoshimoto, Katsutoshi Hori
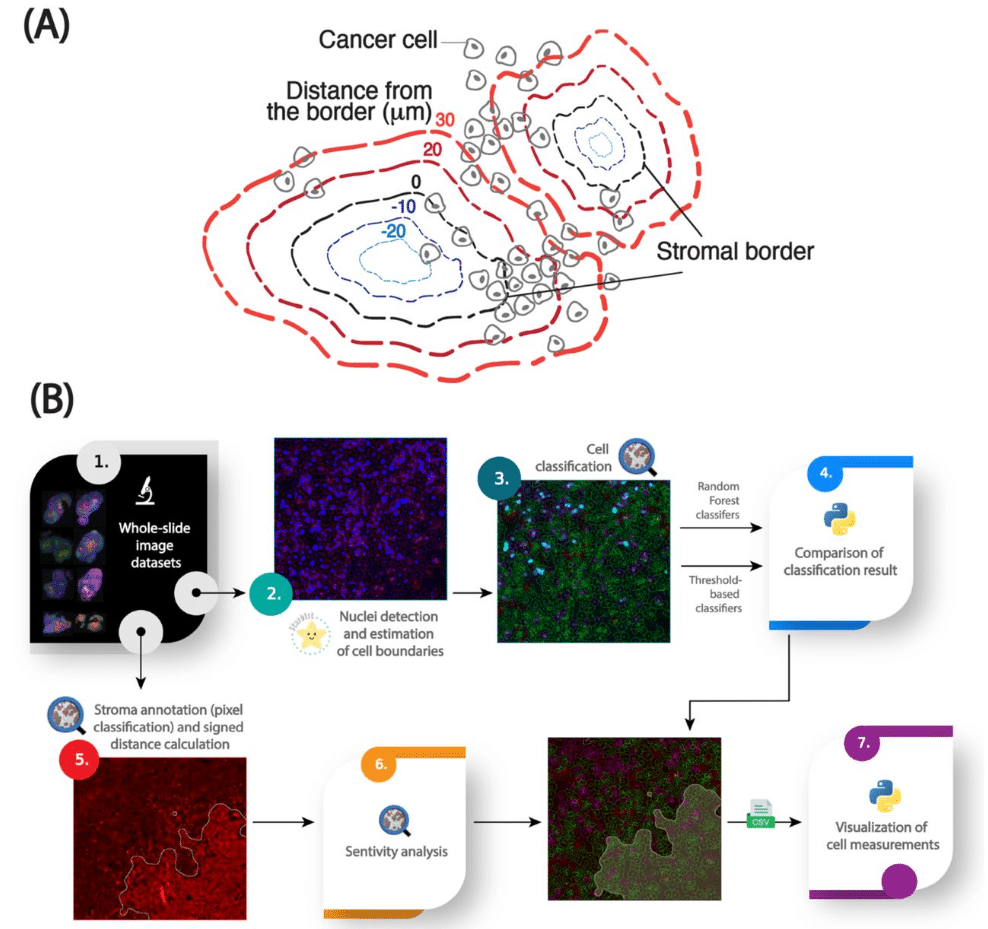
An image analysis pipeline to quantify the spatial distribution of cell markers in stroma-rich tumors
Antoine A. Ruzette, Nina Kozlova, Kayla A Cruz, Taru Muranen, Simon F. Nørrelykke
Streamlining Multiplexed Tissue Image Analysis with PIPΣX: An Integrated Automated Pipeline for Image Processing and EXploration for Diverse Tissue Types
Mariya Mardamshina, Frederic Ballllosera Navarro, Anna Martinez Casals, Christophe Avenel, Carolina Wählby, Emma Lundberg
Spatialproteomics – an interoperable toolbox for analyzing highly multiplexed fluorescence image data
Matthias Meyer-Bender, Harald Voehringer, Christina Schniederjohann, Sarah Koziel, Erin Chung, Ekaterina Popova, Alexander Brobeil, Lisa-Maria Held, Aamir Munir, Scverse Community, Sascha Dietrich, Peter-Martin Bruch, Wolfgang Huber
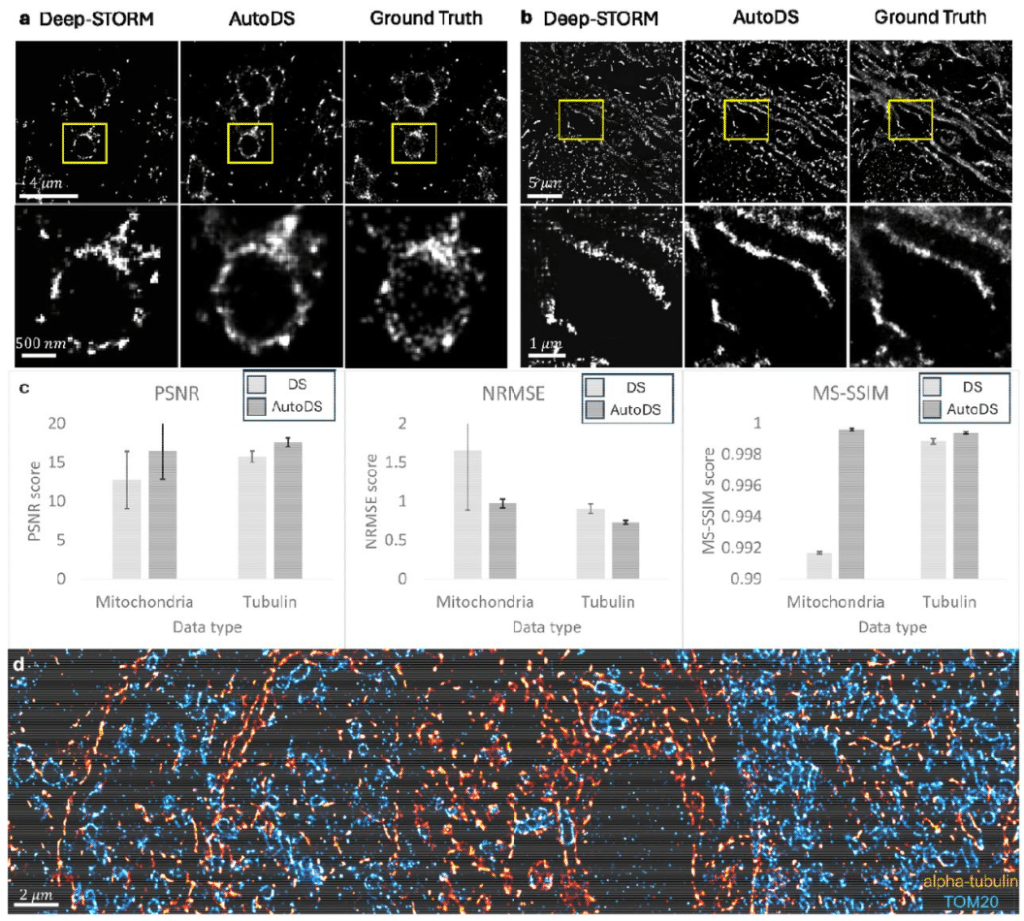
One-click image reconstruction in single-molecule localization microscopy via deep learning
Alon Saguy, Dafei Xiao, Kaarjel K. Narayanasamy, Yuya Nakatani, Anna-Karin Gustavsson, Mike Heilemann, Yoav Shechtman
Fast and accessible morphology-free functional fluorescence imaging analysis
Alejandro Estrada Berlanga, Gabrielle Kang, Amanda Kwok, Thomas Broggini, Jennifer Lawlor, Kishore Kuchibhotla, David Kleinfeld, Gal Mishne, Adam S. Charles
VaMiAnalyzer: An open source, python-based application for analysis of 3D in vitro vasculogenic mimicry assays
Stephen P.G. Moore, Xinyu Zhang, Olivia Chika Jonathan, Anqi Zou, Deborah Lang, Chao Zhang
CABaNe, an automated, high content ImageJ macro for cell and neurite analysis
Nathan Thibieroz, Fabrice Cordelières, Paul Machillot, Akshita Singh, Catherine Picart, Elisa Migliorini
LivecellX: A Scalable Deep Learning Framework for Single-Cell Object-Oriented Analysis in Live-Cell Imaging
Ke Ni, Gaohan Yu, Zhiqian Zheng, Yong Lu, Dante Poe, Yanshuo Chen, Mia Sanborn, Zilin Wang, Shiman Zhou, Xueying Zhan, Weikang Wang, Jianhua Xing
Comparison of QuPath and HALO platforms for analysis of the tumor microenvironment in prostate cancer
wei zhang, Qin Zhou, Jonathan V Nguyen, Erika Egal, Qian Yang, Michael R. Freeman, Siwen Hu-Lieskovan, Gita Suneja, Anna Coghill, Beatrice S Knudsen
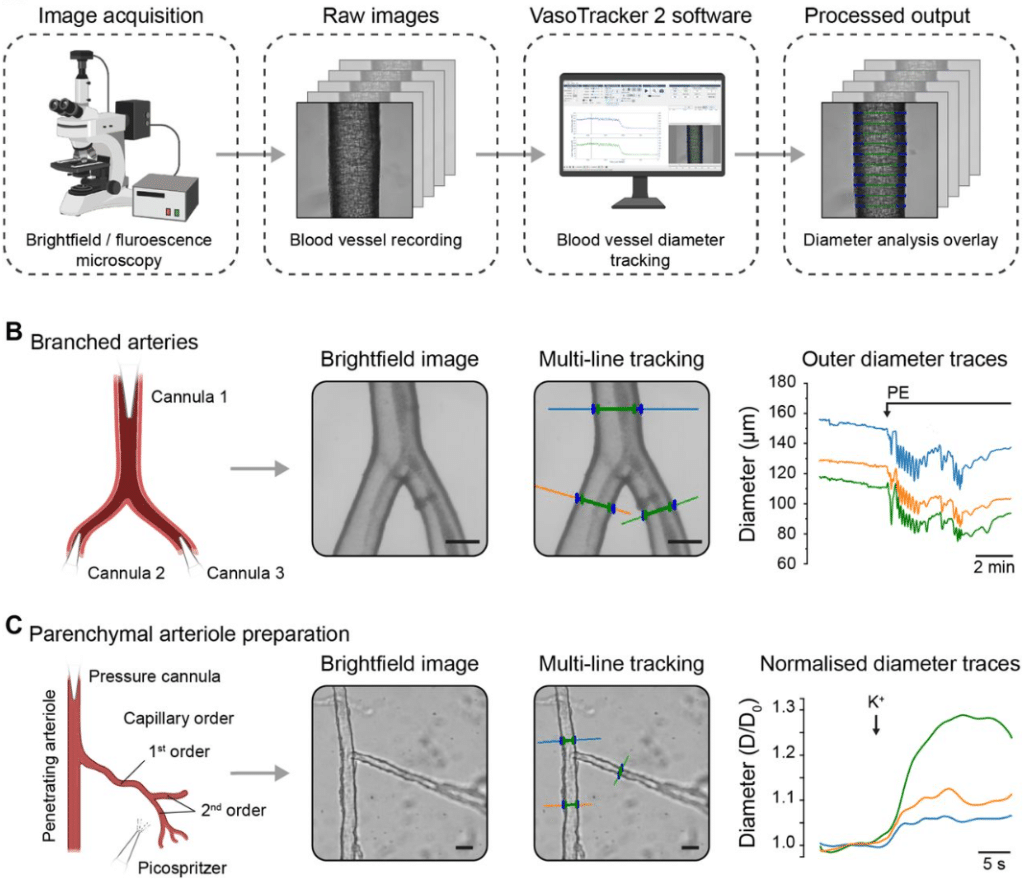
VasoTracker 2: An Open-source Platform for Quantitative Analysis of Vascular Reactivity and Function
Matthew D. Lee, Christopher Osborne, Ross Stevenson, Amy MacDonald, Grace Ebner, Danielle A. Jeffrey, Margaret A. Macdonald, Xun Zhang, Charlotte Buckley, Fabrice Dabertrand, Daniel R Machin, Jason Au, Osama F. Harraz, Nathan Tykocki, John G. McCarron, Calum Wilson
SarcAsM: AI-based multiscale analysis of sarcomere organization and contractility in cardiomyocytes
Daniel Härtter, Lara Hauke, Til Driehorst, Yuxi Long, Guobin Bao, Andreas Primeßnig, Branimir Berečić, Lukas Cyganek, Malte Tiburcy, Christoph F. Schmidt, Wolfram-Hubertus Zimmermann
CellSeg3D: self-supervised 3D cell segmentation for fluorescence microscopy
Cyril Achard, Timokleia Kousi, Markus Frey, Maxime Vidal, Yves Paychère, Colin Hofmann, Asim Iqbal, Sebastien B. Hausmann, Stéphane Pagès, Mackenzie Weygandt Mathis
Contextual High-throughput 3D Volume Electron Microscopy Data Acquisition Using Artificial Intelligence
Tereza Hurník Konečná, Radek Jančík, Daniela Slamková, Bronislav Přibyl, Cveta Tomova, Lolita Rotkina, Tessa Burch-Smith, Sanja Sviben, James A.J. Fitzpatrick, Kirk J. Czymmek
Self-supervised learning enables unbiased patient characterization from multiplexed cancer tissue microscopy images
Gantugs Atarsaikhan, Isabel Mogollon, Katja Välimäki, iCAN, Tuomas Mirtti, Teijo Pellinen, Lassi Paavolainen
Enhancing Biomedical Optical Volumetric Imaging via Self-Supervised Orthogonal Learning
Yuanjie Gu, Yiqun Wang, Ang Xuan, Jianping Wang, Linyi Wang, Lei Zhang, Xiaoran Li, Yao Wu, Jun Zhang, Zhi Lu, Biqin Dong
NuclePhaser: a YOLO-based framework for cell nuclei detection and counting in phase contrast images of arbitrary size with support of fast calibration and testing on specific use cases
Nikita Voloshin, Egor Putlyaev, Elizaveta Chechekhina, Vladimir Usachev, Maxim Karagyaur, Kirill Bozov, Olga Grigorieva, Pyotr Tyurin-Kuzmin, Konstantin Kulebyakin
Simultaneous particle tracking, phase retrieval and point spread function reconstruction
Mohamadreza Fazel, Reza Hoseini, Maryam Mahmoodi, Lance W. Q. Xu, Ayush Saurabh, Zeliha Kilic, Julian Antolin, Kevin L. Scrudders, Douglas Shepherd, Shalini T. Low-Nam, Fang Huang, Steve Pressé
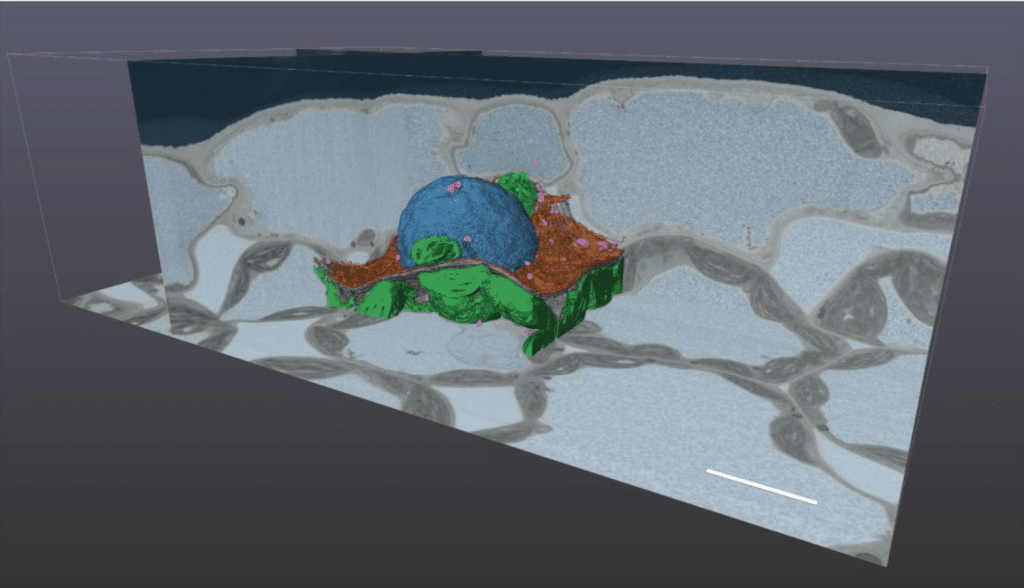
Accurate and fast segmentation of filaments and membranes in micrographs and tomograms with TARDIS
Robert Kiewisz, Gunar Fabig, Will Conway, Jake Johnston, Victor A. Kostyuchenko, Aaron Tan, Cyril Bařinka, Oliver Clarke, Magdalena Magaj, Hossein Yazdkhasti, Francesca Vallese, Shee-Mei Lok, Stefanie Redemann, Thomas Müller-Reichert, Tristan Bepler
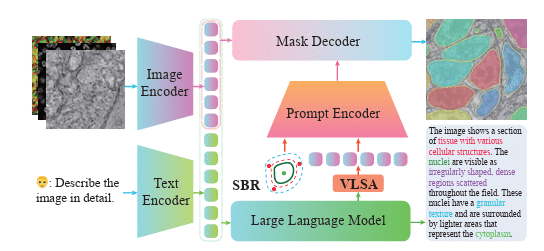
Unifying Segment Anything in Microscopy with Multimodal Large Language Model
Manyu Li, Ruian He, Zixian Zhang, Weimin Tan, Bo Yan
Automated grading and staging of ovarian cancer using deep learning on the transmission optical microscopy bright-field images of thin biopsy tissue samples
Ashmit K Mishra, Mousa Alrubayan, Prabhakar Pradhan
Total Variation-Based Image Decomposition and Denoising for Microscopy Images
Marco Corrias, Giada Franceschi, Michele Riva, Alberto Tampieri, Karin Föttinger, Ulrike Diebold, Thomas Pock, Cesare Franchini
Uni-AIMS: AI-Powered Microscopy Image Analysis
Yanhui Hong, Nan Wang, Zhiyi Xia, Haoyi Tao, Xi Fang, Yiming Li, Jiankun Wang, Peng Jin, Xiaochen Cai, Shengyu Li, Ziqi Chen, Zezhong Zhang, Guolin Ke, Linfeng Zhang
Cascade Detector Analysis and Application to Biomedical Microscopy
Thomas L. Athey, Shashata Sawmya, Nir Shavit
SAM4EM: Efficient memory-based two stage prompt-free segment anything model adapter for complex 3D neuroscience electron microscopy stacks
Uzair Shah, Marco Agus, Daniya Boges, Vanessa Chiappini, Mahmood Alzubaidi, Jens Schneider, Markus Hadwiger, Pierre J. Magistretti, Mowafa Househ, Corrado Calı
DiffKillR: Killing and Recreating Diffeomorphisms for Cell Annotation in Dense Microscopy Images
Chen Liu, Danqi Liao, Alejandro Parada-Mayorga, Alejandro Ribeiro, Marcello DiStasio, Smita Krishnaswamy
BigReg: An Efficient Registration Pipeline for High-Resolution X-Ray and Light-Sheet Fluorescence Microscopy
Siyuan Mei, Fuxin Fan, Mareike Thies, Mingxuan Gu, Fabian Wagner, Oliver Aust, Ina Erceg, Zeynab Mirzaei, Georgiana Neag, Yipeng Sun, Yixing Huang, Andreas Maier
BioVFM-21M: Benchmarking and Scaling Self-Supervised Vision Foundation Models for Biomedical Image Analysis
Jiarun Liu, Hong-Yu Zhou, Weijian Huang, Hao Yang, Dongning Song, Tao Tan, Yong Liang, Shanshan Wang


 (No Ratings Yet)
(No Ratings Yet)