Microscopy preprints – bioimage analysis
Posted by FocalPlane, on 14 November 2025
Here is a curated selection of preprints published recently. In this post, we focus specifically on bioimage analysis and data management.
Lightweight open-source fine-tuning of SAM2 enables domain-specific microscopy segmentation
Etash Bhat, Sharvaa Selvan, Sonia Okekenwa, Zareh Dechkounian, Vivian Lin, Mayu Nakano, Mitul Saha, Yuyu Song
Lessons learned from a Kaggle challenge for particle picking in cryo-electron tomography
Ariana Peck, Joshua Hutchings, Jonathan Schwartz, Yue Yu, Utz H. Ermel, Saugat Kandel, Dari Kimanius, Zhuowen Zhao, Shawn Zheng, Brendan Artley, David List, Sergio A. Silva Jr., Walter Reade, Jeremy Asuncion, Kira Evans, Jessica Gadling, Kandarp Khandwala, Suzette McCanny, Dannielle G. McCarthy, Jun Xi Ni, Janeece Pourroy, Manasa Venkatakrishnan, Zun Shi Wang, David A. Agard, Clinton S. Potter, Bridget Carragher, Kyle Harrington, Mohammadreza Paraan
Deep-learning deconvolution and segmentation of fluorescent membranes for high-precision bacterial cell-size profiling
Octavio Reyes-Matte, Carsten Fortmann-Grote, Beate Gericke, Nadja Hüttmann, Nikola Ojkic, Javier Lopez-Garrido
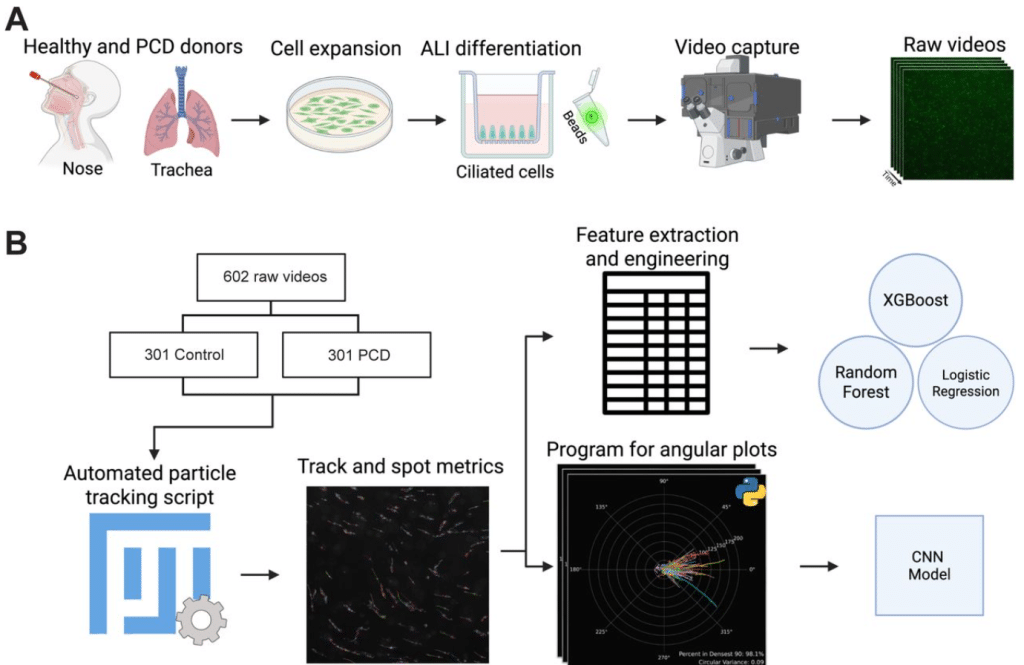
Machine Learning Analysis of Cilia-Driven Particle Transport Distinguishes Primary Ciliary Dyskinesia Cilia from Normal Cilia
Nicholas Hadas, Huihui Xu, Shambhawee Neupane, Wang K Twan, Ahmed Elgamal, Amjad Horani
Combining FRET and super-resolution microscopy reveals kinase activation and mitochondrial activity at the nanoscale
Nicolas Y. Jolivet, Pierre-Jean Desmaison, Xavier Pinson, Olivier Delalande, Giulia Bertolin
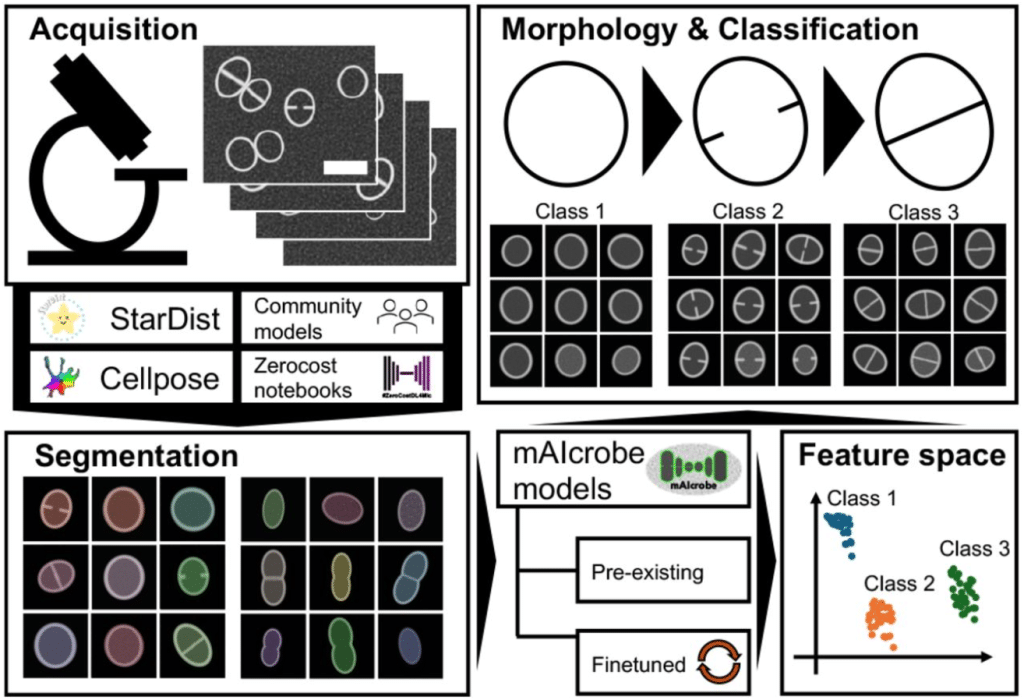
mAIcrobe: an open-source framework for high-throughput bacterial image analysis
António D. Brito, Dominik Alwardt, Beatriz de P. Mariz, Sérgio R. Filipe, Mariana G. Pinho, Bruno M. Saraiva, Ricardo Henriques
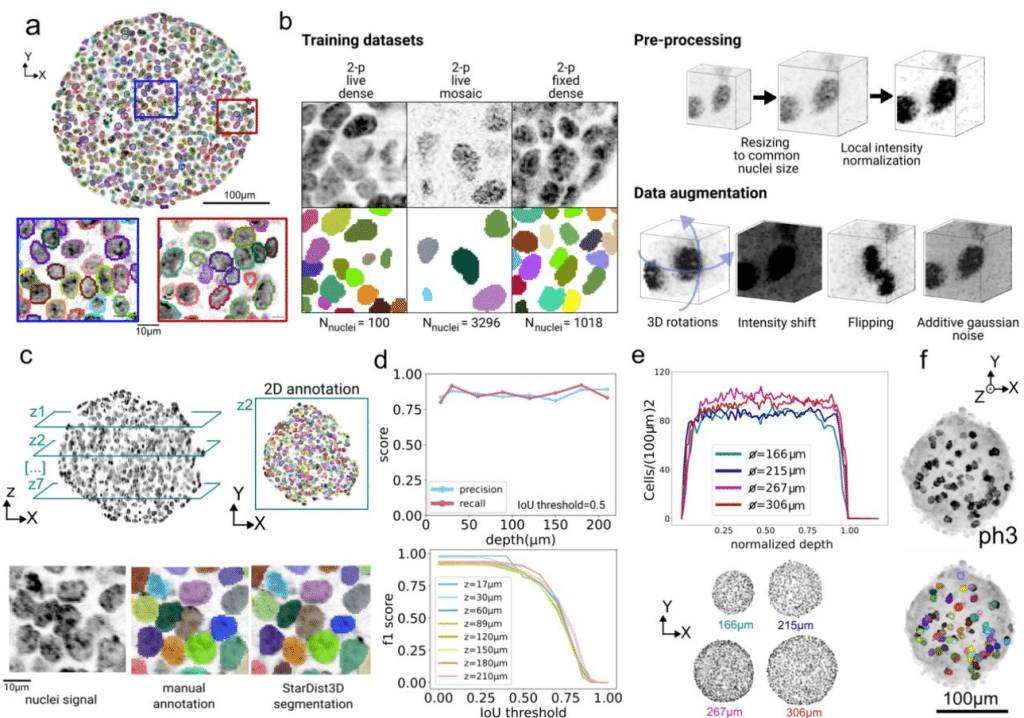
A quantitative pipeline for whole-mount deep imaging and analysis of multi-layered organoids across scales
Alice Gros, Jules Vanaret, Valentin Dunsing-Eichenauer, Agathe Rostan, Philippe Roudot, Pierre-François Lenne, Léo Guignard, Sham Tlili
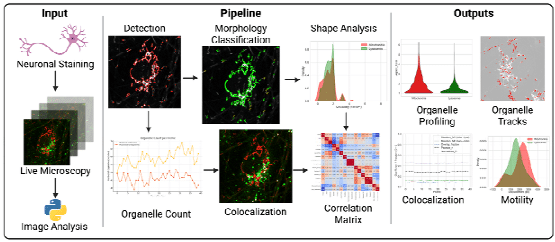
AutoMorphoTrack: A modular framework for quantitative analysis of organelle morphology, motility, and interactions at single-cell resolution
Armin Bayati, Jackson G. Schumacher, Xiqun Chen
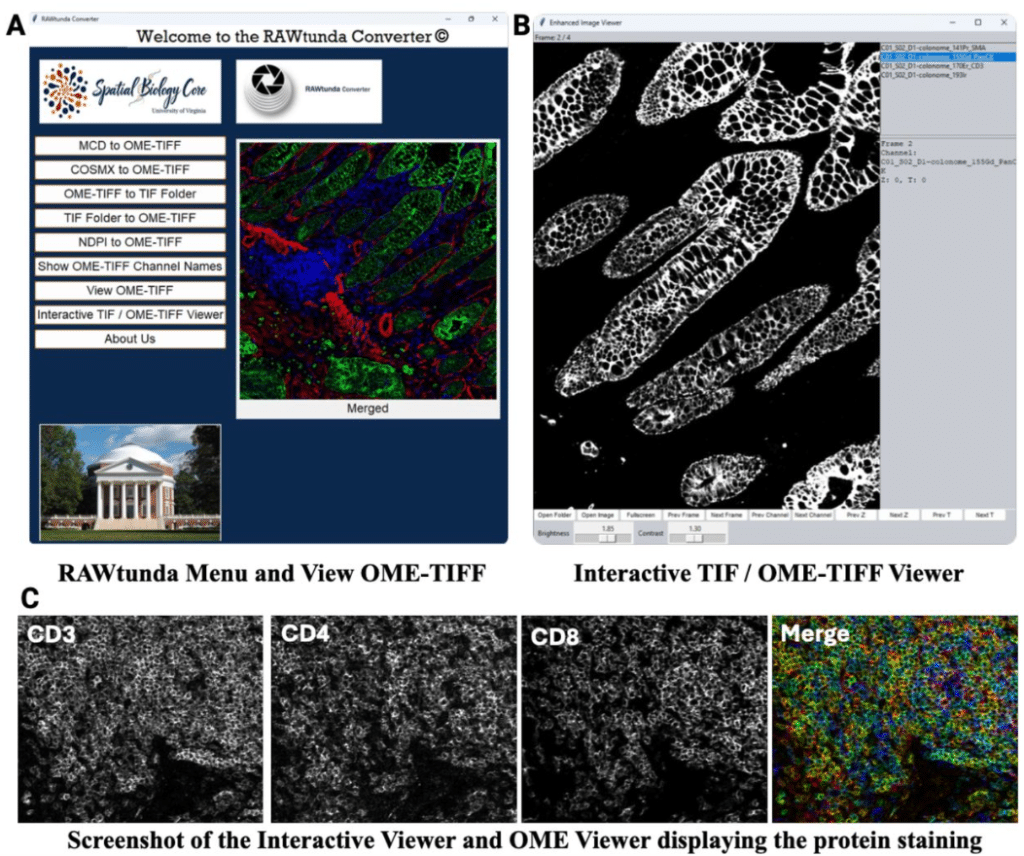
RAWtunda: a Tool to Convert a Multi-Channel Raw Image into TIFF and OME-TIFF Formats
Natalia M. Dworak, Maxwell Cooper, Jay W. Fox, Ana K. de Oliveira
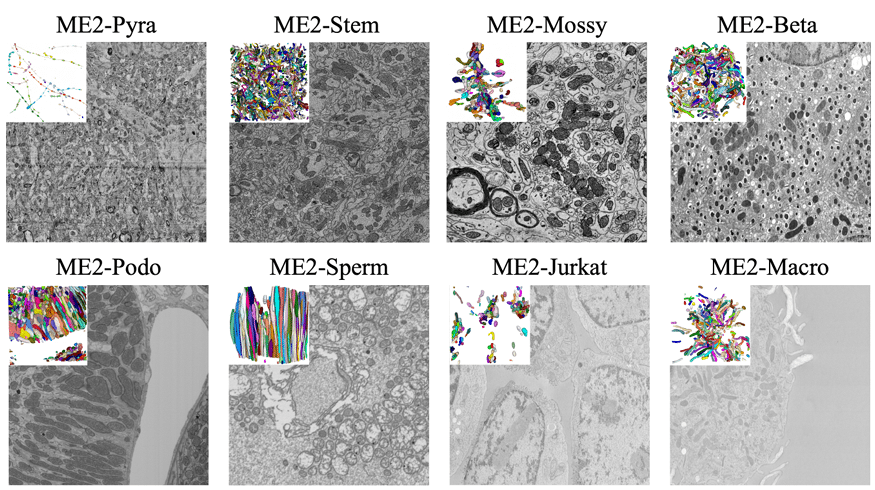
MitoEM 2.0 – A Benchmark for Challenging 3D Mitochondria Instance Segmentation from EM Images
Peng Liu, Boyu Shen, Liyuan Liu, Qiong Wang, Shulin Zhang, Abhishek Bhardwaj, Ignacio Arganda-Carreras, Kedar Narayan, Donglai Wei
Automated filtering of particle images in single particle cryoEM
Sony Malhotra, Daniel Hatton, Samuel Jackson, Matthew Iadanza, Agnel Praveen Joseph, Colin Palmer, Jeyan Thiyagalingam, Tom Burnley, Yuriy Chaban
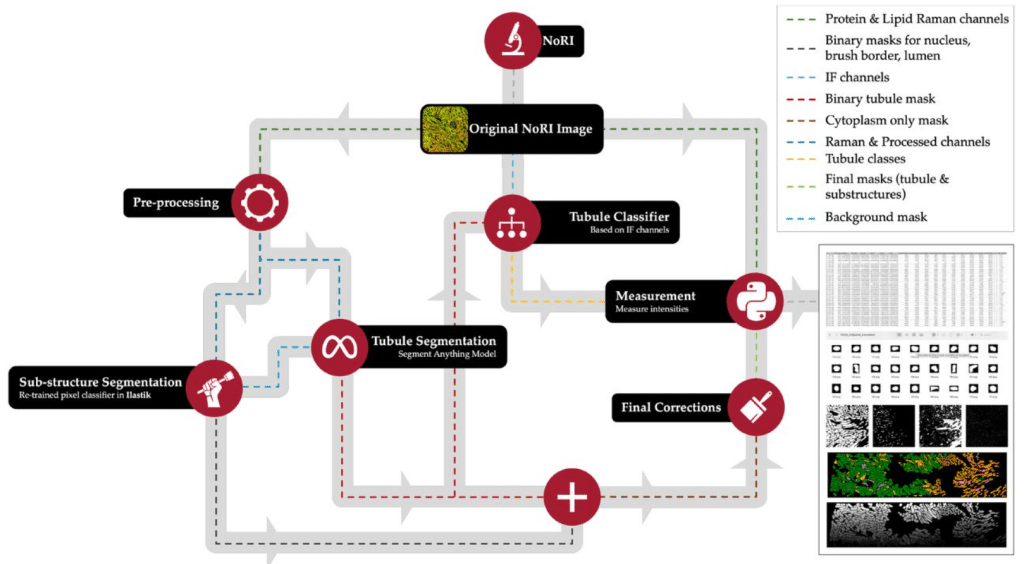
Kidney Tissue Characterization using Normalized Raman Imaging and Segment-Anything
Ranit Karmakar, William V. Trim, Marc Kirschner, Simon F. Nørrelykke
MDMR: Balancing Diversity and Redundancy for Annotation-Efficient Fine-Tuning of Pretrained Cell Segmentation Models
Eiram Mahera Sheikh, Alaa Tharwat, Constanze Schwan, Wolfram Schenck
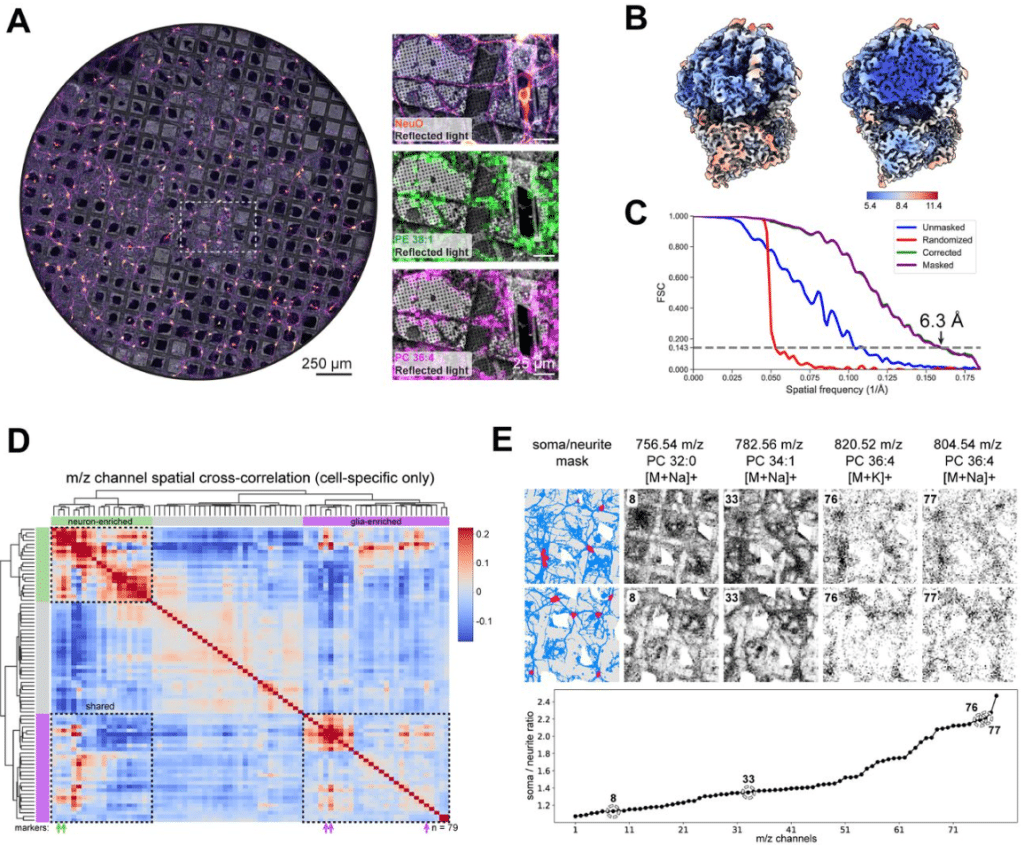
Correlative MS Imaging for in situ cryo-ET
Andre Schwarz, Jakob Meier-Credo, Arne Behrens, Jürgen Reichert, Sonja Welsch, Lea Dietrich, Erin M. Schuman, Julian D. Langer
Leonardo: a toolset to correct sample-induced artifacts in light sheet microscopy images
Yu Liu, Gesine F. Müller, Lennart Kowitz, Tomáš Chobola, Kurt Weiss, Paul Maier, Jie Luo, Malte Roeßing, Martin Stenzel, Anika Grüneboom, Johannes Paetzold, Ali Ertürk, Nassir Navab, Carsten Marr, Jianxun Chen, Jan Huisken, Tingying Peng
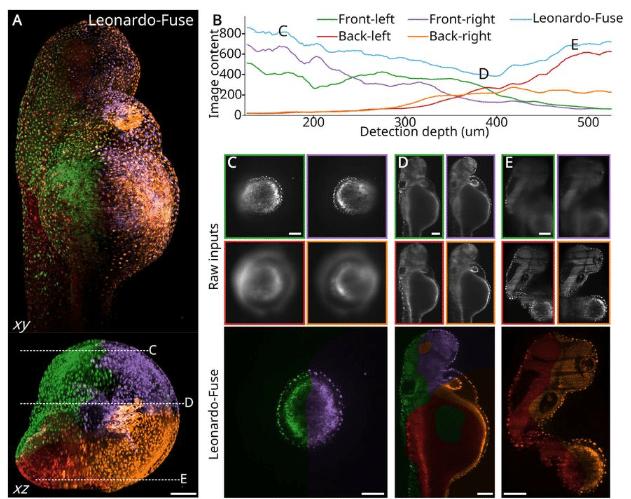
Image Analysis for Non-Neoplastic Kidney Disease: Utilizing Morphological Segmentation to Improve Quantification of Interstitial Fibrosis
Nazanin Mola, Maya Maya Barbosa Silva, Sabine Leh, Hrafn Weishaupt
An automated image analysis pipeline for wide-field optical redox imaging of patient-derived cancer organoids
Angela Hsu, Kayvan Samimi, Amani A. Gillette, Shirsa Udgata, Alexa E. Schmitz, Wenxuan Zhao, Dustin A. Deming, Melissa C. Skala
CellMAPS: A no-code model-based customisable multiplex image analysis workflow
Éanna Fennell, Colm Brandon, Steve Boßelmann, Stephen E. Ryan, Amandeep Singh, Aoife Hennessy, Aisling M. Ross, Ciara I. Leahy, Matthew R. Pugh, Nadezhda Nikulina, Oliver Braubach, Gerald Niedobitek, Graham S. Taylor, Tiziana Margaria, Paul G. Murray
Innovative 3D-image analysis of cerebellar vascularization highlights angiogenic gene dysregulations in a murine model of apnea of prematurity
A. Rodriguez-Duboc, C. Racine, M. Basille-Dugay, D. Vaudry, B. Gonzalez, D. Burel
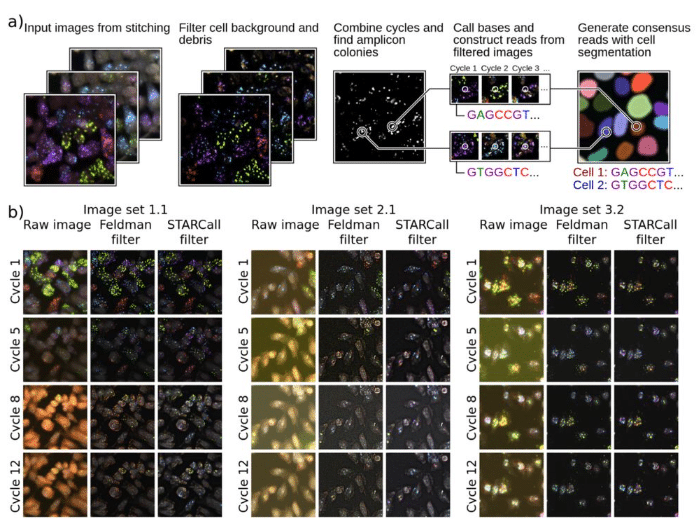
STARCall integrates image stitching, alignment, and read calling to enable scalable analysis of in situ sequencing data
Nicholas J. Bradley, Sriram Pendyala, Katie Partington, Douglas M. Fowler
cryoJAX: A Cryo-electron Microscopy Image Simulation Library In JAX
Michael J. O’Brien, David Silva-Sánchez, Geoffrey Woollard, Kwanghwi Je, Sonya M. Hanson, Daniel J. Needleman, Pilar Cossio, Erik Henning Thiede, Miro A. Astore
Twist and Scout: Analysis and Curation of Particles in Cryo-Electron Tomography Using TANGO
Markus Schreiber, Beata TuroňováTwist and Scout: Analysis and Curation of Particles in Cryo-Electron Tomography Using TANGO
Markus Schreiber, Beata Turoňová
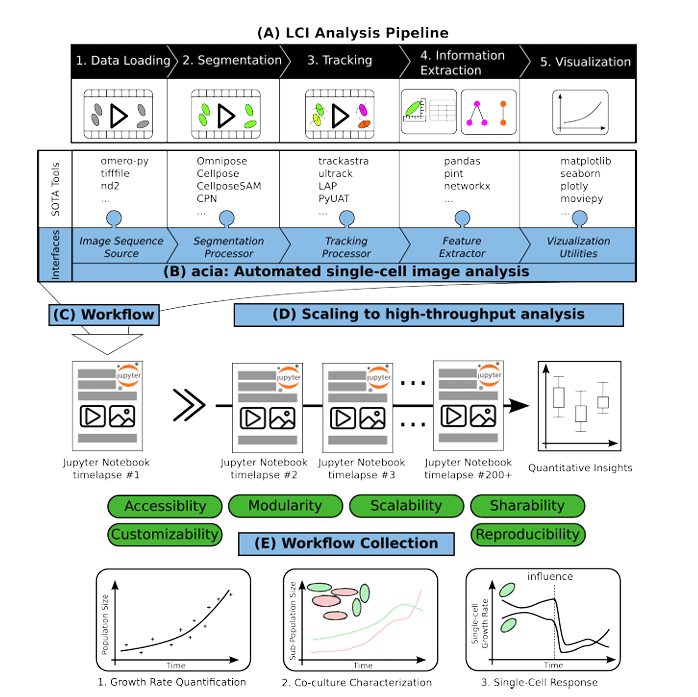
acia-workflows: Automated Single-cell Imaging Analysis for Scalable and Deep Learning-based Live-cell Imaging Analysis Workflows
Johannes Seiffarth, Keitaro Kasahara, Michelle Bund, Benita Lückel, Richard D. Paul, Matthias Pesch, Lennart Witting, Michael Bott, Dietrich Kohlheyer, Katharina Nöh


 (No Ratings Yet)
(No Ratings Yet)