OMERO Outputs: Insights from the 4th BioHackathon 2025
Posted by Lea Kabjesz, on 17 December 2025
Hi all! Today I would like to share my experience participating in my first BioHackathon and point you to some new exciting features coming soon to the bioimaging software OMERO. If you are interested in participating in a hackathon yourself, but don’t know what to expect or where to start, I have prepared some insights and helpful pointers to get you started.
But let’s go back to the beginning. The 4th BioHackathon took place from December 1-5th in Walsrode, Germany. More than 80 participants came together to work on 11 different projects. The whole event was organized by the German Network for Bioinformatics Infrastructure – or short: de.NBI. Most of you will probably be more familiar with its European network, called ELIXIR. The goal of both initiatives is to support life science research by providing sustainable infrastructure, tools, standards, and training.

Group photo of participants at the 4th BioHackathon Germany, 2025.
The OMERO project
I was part of a project focusing on the OMERO software. To all my biology-friends out there working with microscopy data: I highly encourage you to check this out! OMERO is a server-based platform which you can use to store, share, visualize, and annotate imaging data. Beyond storage, OMERO offers plugins and integrations with many commonly used analysis tools such as napari, Fiji/ImageJ, and Jupyter Notebooks, enabling you to build analysis workflows.
Another really nice thing about OMERO is that it’s open-source! This means that developers from the community are continuously working on the code to make the software more user-friendly. Users can actively contribute and shape the software by reporting bugs, requesting features, or even contributing code themselves. The main place where this happens is on GitHub. Simply send us a message (so called “issue”). If you are new to GitHub, don’t worry, my colleague Mara Lampert wrote a very nice blogpost explaining how to create an issue.
GitHub. GitHub is the world’s largest developer platform for hosting and managing code. It is especially helpful for collaborative work and version control.
A week of hacking (and more…)
So what happened during our week? Let me tell you: quite a lot! Besides hacking, there were many social activities such as a pub-quiz night, board games, bowling, and dancing. On day 1, we all met in a large seminar hall, where the project leads presented their projects and goals. The topics were incredibly diverse, ranging from agriculture and biomedicine to learning materials, data standards, and many more …. You can find an overview of all topics here. What was especially interesting to me was that – contrary to my expectations – many projects were working more on a conceptual level and were less programming-heavy. While, of course, every hackathon and every project is different, I want to encourage you to join a hackathon if you get the chance – even if you have NO coding experience at all! All projects were happy to reach members interested in their topic (even if coming from a completely different background). In the end, it’s all about discussion, getting new ideas, or just reaching potential users.

Project Lead Tom Boissonnet presenting the OMERO project.
After the project pitches, we all split into separate rooms dedicated to each project. And then it began: a never-ending cycle of hacking, coffee, hacking, coffee, … (and a lot of food). Depending on the task and individual preferences, our team worked in different ways: small groups, pair-programming, or individually. Since many people in our group come from different OMERO plugins, this setting was a great opportunity to finally get together and tackle broader issues – features that need to be implemented across multiple plugins, or design decisions that benefit from multiple perspectives. For me personally, being a newbie to the OMERO codebase, it was incredibly helpful to be able to quickly ask someone to look over my code before getting stuck.

The OMERO on-site and online project team.
Our Outputs
During the hackathon, I tackled an issue by Anna Hamacher, who asked for better support for high-content screening data. We added options that allow users to automatically label wells, plates, screens, and runs directly in OMERO.figure – eliminating the need to tediously go back to the metadata and copy-paste labels when creating scientific figures. I also addressed an issue for exporting images in PNG format at lower resolution – a feature request raised by the community for presentations and publications. Of course, many thanks to all my team members, especially Tom Boissonnet, for their help whenever I had questions during implementation!
Overall, I felt that the collaborative environment really pushed all of us to be productive. Our team tackled over ten different features, leading to many new improvements that will be launched in OMERO soon. You can already access some features, while others are still going through testing before they are publicly released. Here is a small visual sneak peek at a subset of what we developed during the week:
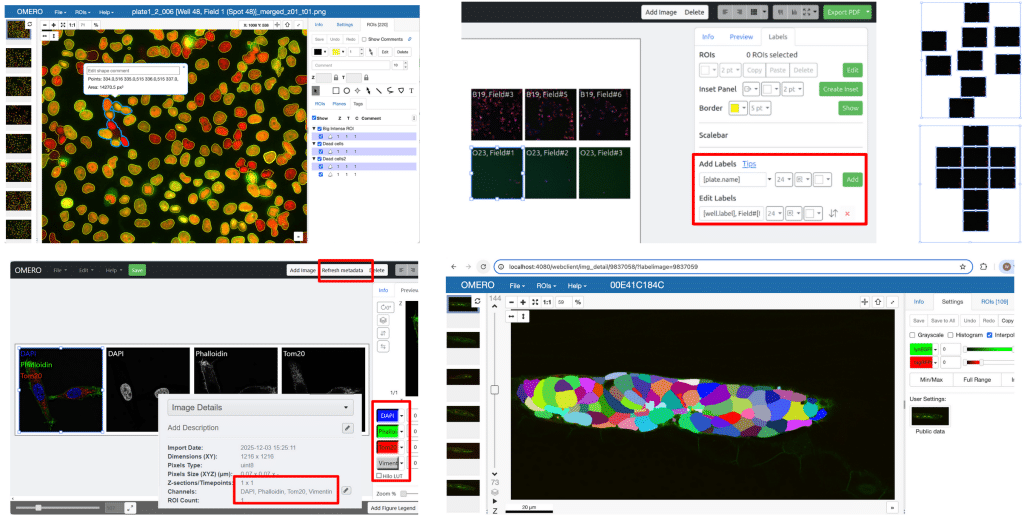
Collage showcasing a selection of new OMERO features developed during the hackathon.
- ROI tagging in OMERO.iviewer and OMERO.parade2
- Plate labeling and lower resolution export in OMERO.figure
- Align to grid option in OMERO.figure
- Refresh metadata button in OMERO.figure
- OMERO collections (RFC-8) in napari-omero and iviewer to load labels, annotations, and masks
- Grid extension in OMERO.iviewer for palynological data
- Galaxy OMERO-suite release to simplify uploading data from OMERO
- OMERO.parade2 crossfilter for plotting tabular data
- OME-NGFF → 3D Slicer integration
- Jupyter Notebooks for training/finetuning using data from OMERO
Learn more about these features at the FDM-Werkstatt 2026 in Düsseldorf, Germany! 🔬
Further Reading
- Project Presentation Slides: https://zenodo.org/records/17856704
- Project Chat and Outputs: https://imagesc.zulipchat.com/#narrow/channel/549193-.5B2025-12.5D-de.2ENBI-BioHackathon-Walsrode
Acknowledgments
This Blogpost acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under the National Research Data Infrastructure – NFDI 46/1 – 501864659.
Many thanks to de.NBI and its entire organizing-team for putting together this rewarding event. We had an exciting (albeit also exhausting 😉) week of hacking, collaborating, and social activities!
Reusing this material
This blog post is open-access. Reuse of content is permitted under the terms of the CC BY 4.0 license, unless stated otherwise.
Feedback welcome
I’m interested in the day-to-day challenges life scientists face when working with imaging data. My goal is to support the community in this field – whether through developing practical tools or creating training materials with blog posts like these. Feel free to reach out if anything is unclear or if you’d simply like to connect with ideas for tool developments or collaboration! 🤝


 (No Ratings Yet)
(No Ratings Yet)